Genome-wide detection of spontaneous chromosomal rearrangements in bacteria
- PMID: 22880062
- PMCID: PMC3411829
- DOI: 10.1371/journal.pone.0042639
Genome-wide detection of spontaneous chromosomal rearrangements in bacteria
Abstract
Genome rearrangements have important effects on bacterial phenotypes and influence the evolution of bacterial genomes. Conventional strategies for characterizing rearrangements in bacterial genomes rely on comparisons of sequenced genomes from related species. However, the spectra of spontaneous rearrangements in supposedly homogenous and clonal bacterial populations are still poorly characterized. Here we used 454 pyrosequencing technology and a 'split mapping' computational method to identify unique junction sequences caused by spontaneous genome rearrangements in chemostat cultures of Salmonella enterica Var. Typhimurium LT2. We confirmed 22 unique junction sequences with a junction microhomology more than 10 bp and this led to an estimation of 51 true junction sequences, of which 28, 12 and 11 were likely to be formed by deletion, duplication and inversion events, respectively. All experimentally confirmed rearrangements had short inverted (inversions) or direct (deletions and duplications) homologous repeat sequences at the endpoints. This study demonstrates the feasibility of genome wide characterization of spontaneous genome rearrangements in bacteria and the very high steady-state frequency (20-40%) of rearrangements in bacterial populations.
Conflict of interest statement
Figures

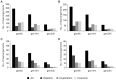


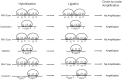

References
-
- Suyama M, Bork P (2001) Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet 17: 10–13. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

