p100 Deficiency is insufficient for full activation of the alternative NF-κB pathway: TNF cooperates with p52-RelB in target gene transcription
- PMID: 22880094
- PMCID: PMC3412832
- DOI: 10.1371/journal.pone.0042741
p100 Deficiency is insufficient for full activation of the alternative NF-κB pathway: TNF cooperates with p52-RelB in target gene transcription
Abstract
Background: Constitutive activation of the alternative NF-κB pathway leads to marginal zone B cell expansion and disorganized spleen microarchitecture. Furthermore, uncontrolled alternative NF-κB signaling may result in the development and progression of cancer. Here, we focused on the question how does the constitutive alternative NF-κB signaling exert its effects in these malignant processes.
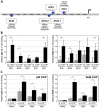
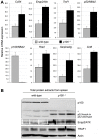
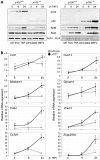
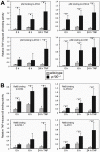
Methodology/principal findings: To explore the consequences of unrestricted alternative NF-κB activation on genome-wide transcription, we compared gene expression profiles of wild-type and NF-κB2/p100-deficient (p100(-/-)) primary mouse embryonic fibroblasts (MEFs) and spleens. Microarray experiments revealed only 73 differentially regulated genes in p100(-/-) vs. wild-type MEFs. Chromatin immunoprecipitation (ChIP) assays showed in p100(-/-) MEFs direct binding of p52 and RelB to the promoter of the Enpp2 gene encoding ENPP2/Autotaxin, a protein with an important role in lymphocyte homing and cell migration. Gene ontology analysis revealed upregulation of genes with anti-apoptotic/proliferative activity (Enpp2/Atx, Serpina3g, Traf1, Rrad), chemotactic/locomotory activity (Enpp2/Atx, Ccl8), and lymphocyte homing activity (Enpp2/Atx, Cd34). Most importantly, biochemical and gene expression analyses of MEFs and spleen, respectively, indicated a marked crosstalk between classical and alternative NF-κB pathways.
Conclusions/significance: Our results show that p100 deficiency alone was insufficient for full induction of genes regulated by the alternative NF-κB pathway. Moreover, alternative NF-κB signaling strongly synergized both in vitro and in vivo with classical NF-κB activation, thereby extending the number of genes under the control of the p100 inhibitor of the alternative NF-κB signaling pathway.
Conflict of interest statement
Figures

Similar articles
-
Immune Differentiation Regulator p100 Tunes NF-κB Responses to TNF.Front Immunol. 2019 May 7;10:997. doi: 10.3389/fimmu.2019.00997. eCollection 2019. Front Immunol. 2019. PMID: 31134075 Free PMC article.
-
Regulation of p53 and Rb links the alternative NF-κB pathway to EZH2 expression and cell senescence.PLoS Genet. 2014 Sep 25;10(9):e1004642. doi: 10.1371/journal.pgen.1004642. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25255445 Free PMC article.
-
NF-κB2/p100 deficiency impairs immune responses to T-cell-independent type 2 antigens.Eur J Immunol. 2014 Mar;44(3):662-72. doi: 10.1002/eji.201343484. Epub 2013 Dec 16. Eur J Immunol. 2014. PMID: 24242887
-
The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development.Biochem Pharmacol. 2006 Oct 30;72(9):1161-79. doi: 10.1016/j.bcp.2006.08.007. Epub 2006 Sep 12. Biochem Pharmacol. 2006. PMID: 16970925 Review.
-
Alternative pathways of NF-kappaB activation: a double-edged sword in health and disease.Cytokine Growth Factor Rev. 2006 Aug;17(4):281-93. doi: 10.1016/j.cytogfr.2006.04.005. Epub 2006 Jun 21. Cytokine Growth Factor Rev. 2006. PMID: 16793322 Review.
Cited by
-
Friend or Foe: Regulation, Downstream Effectors of RRAD in Cancer.Biomolecules. 2023 Mar 5;13(3):477. doi: 10.3390/biom13030477. Biomolecules. 2023. PMID: 36979412 Free PMC article. Review.
-
Tumor necrosis factor-α attenuates starvation-induced apoptosis through upregulation of ferritin heavy chain in hepatocellular carcinoma cells.BMC Cancer. 2013 Sep 25;13:438. doi: 10.1186/1471-2407-13-438. BMC Cancer. 2013. PMID: 24066693 Free PMC article.
-
Autotaxin in Pathophysiology and Pulmonary Fibrosis.Front Med (Lausanne). 2018 Jun 13;5:180. doi: 10.3389/fmed.2018.00180. eCollection 2018. Front Med (Lausanne). 2018. PMID: 29951481 Free PMC article. Review.
-
The Alternative NF-κB Pathway in Regulatory T Cell Homeostasis and Suppressive Function.J Immunol. 2018 Apr 1;200(7):2362-2371. doi: 10.4049/jimmunol.1800042. Epub 2018 Feb 19. J Immunol. 2018. PMID: 29459403 Free PMC article.
-
Investigating Natural Product Inhibitors of IKKα: Insights from Integrative In Silico and Experimental Validation.Molecules. 2025 May 2;30(9):2025. doi: 10.3390/molecules30092025. Molecules. 2025. PMID: 40363830 Free PMC article.
References
-
- Ghosh S, Karin M (2002) Missing pieces in the NF-kappaB puzzle. Cell 109 Suppl: S81–96. - PubMed
-
- Hayden MS, Ghosh S (2004) Signaling to NF-kappaB. Genes Dev 18: 2195–2224. - PubMed
-
- Hayden MS, Ghosh S (2008) Shared principles in NF-kappaB signaling. Cell 132: 344–362. - PubMed
-
- Hayden MS, West AP, Ghosh S (2006) NF-kappaB and the immune response. Oncogene 25: 6758–6780. - PubMed
-
- Vallabhapurapu S, Karin M (2009) Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27: 693–733. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

