Genomic patterns of homozygosity in worldwide human populations
- PMID: 22883143
- PMCID: PMC3415543
- DOI: 10.1016/j.ajhg.2012.06.014
Genomic patterns of homozygosity in worldwide human populations
Abstract
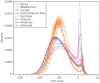
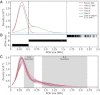
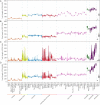
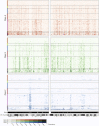
Genome-wide patterns of homozygosity runs and their variation across individuals provide a valuable and often untapped resource for studying human genetic diversity and evolutionary history. Using genotype data at 577,489 autosomal SNPs, we employed a likelihood-based approach to identify runs of homozygosity (ROH) in 1,839 individuals representing 64 worldwide populations, classifying them by length into three classes-short, intermediate, and long-with a model-based clustering algorithm. For each class, the number and total length of ROH per individual show considerable variation across individuals and populations. The total lengths of short and intermediate ROH per individual increase with the distance of a population from East Africa, in agreement with similar patterns previously observed for locus-wise homozygosity and linkage disequilibrium. By contrast, total lengths of long ROH show large interindividual variations that probably reflect recent inbreeding patterns, with higher values occurring more often in populations with known high frequencies of consanguineous unions. Across the genome, distributions of ROH are not uniform, and they have distinctive continental patterns. ROH frequencies across the genome are correlated with local genomic variables such as recombination rate, as well as with signals of recent positive selection. In addition, long ROH are more frequent in genomic regions harboring genes associated with autosomal-dominant diseases than in regions not implicated in Mendelian diseases. These results provide insight into the way in which homozygosity patterns are produced, and they generate baseline homozygosity patterns that can be used to aid homozygosity mapping of genes associated with recessive diseases.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Darwin C.R. John Murray; London, UK: 1876. The effects of cross and self fertilization in the vegetable kingdom.
-
- Garrod A.E. The incidence of alkaptonuria: a study in chemical individuality. Lancet. 1902;160:1616–1620.
-
- Mendel, G. (1866). Versuche über Pflanzenhybriden. Verhandlungen des naturforschenden Vereines in Brünn, 4, 3–47.
-
- Li L.H., Ho S.F., Chen C.H., Wei C.Y., Wong W.C., Li L.Y., Hung S.I., Chung W.H., Pan W.H., Lee M.T. Long contiguous stretches of homozygosity in the human genome. Hum. Mutat. 2006;27:1115–1121. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

