Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137
- PMID: 22883350
- PMCID: PMC4104606
- DOI: 10.1016/j.schres.2012.06.038
Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137
Abstract
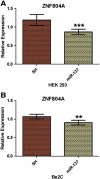
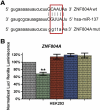
MicroRNAs (miRNAs) are small non-coding RNAs that mainly function as negative regulators of gene expression (Lai, 2002) and have been shown to be involved in schizophrenia etiology through genetic and expression studies (Burmistrova et al., 2007; Hansen et al., 2007a; Perkins et al., 2007; Beveridge et al., 2010; Kim et al., 2010). In a mega analysis of genome-wide association study (GWAS) of schizophrenia (SZ) and bipolar disorders (BP), a polymorphism (rs1625579) located in the primary transcript of a miRNA gene, hsa-miR-137, was reported to be strongly associated with SZ. Four SZ loci (CACNA1C, TCF4, CSMD1, C10orf26) achieving genome-wide significance in the same study were predicted and later experimentally validated (Kwon et al., 2011) as hsa-miR-137 targets. Here, using in silico, cellular and luciferase based approaches we also provide evidence that another well replicated candidate schizophrenia gene, ZNF804A, is also target for hsa-miR-137.
Published by Elsevier B.V.
Figures
Similar articles
-
Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex.J Psychiatr Res. 2013 Sep;47(9):1215-21. doi: 10.1016/j.jpsychires.2013.05.021. Epub 2013 Jun 17. J Psychiatr Res. 2013. PMID: 23786914 Free PMC article.
-
Replication of GWAS identified miR-137 and its target gene polymorphisms in Schizophrenia of South Indian population and meta-analysis with Psychiatric Genomics Consortium.Schizophr Res. 2018 Sep;199:189-194. doi: 10.1016/j.schres.2018.03.028. Epub 2018 Mar 26. Schizophr Res. 2018. PMID: 29599094
-
The regulation of gene expression involved in TGF-β signaling by ZNF804A, a risk gene for schizophrenia.Schizophr Res. 2013 May;146(1-3):273-8. doi: 10.1016/j.schres.2013.01.026. Epub 2013 Feb 22. Schizophr Res. 2013. PMID: 23434502
-
How might ZNF804A variants influence risk for schizophrenia and bipolar disorder? A literature review, synthesis, and bioinformatic analysis.Am J Med Genet B Neuropsychiatr Genet. 2014 Jan;165B(1):28-40. doi: 10.1002/ajmg.b.32207. Epub 2013 Oct 4. Am J Med Genet B Neuropsychiatr Genet. 2014. PMID: 24123948 Review.
-
Bioinformatic analyses and conceptual synthesis of evidence linking ZNF804A to risk for schizophrenia and bipolar disorder.Am J Med Genet B Neuropsychiatr Genet. 2015 Jan;168B(1):14-35. doi: 10.1002/ajmg.b.32284. Am J Med Genet B Neuropsychiatr Genet. 2015. PMID: 25522715 Review.
Cited by
-
Neurodevelopmental concepts of schizophrenia in the genome-wide association era: AKT/mTOR signaling as a pathological mediator of genetic and environmental programming during development.Schizophr Res. 2020 Mar;217:95-104. doi: 10.1016/j.schres.2019.08.036. Epub 2019 Sep 12. Schizophr Res. 2020. PMID: 31522868 Free PMC article.
-
MicroRNA-137 regulates a glucocorticoid receptor-dependent signalling network: implications for the etiology of schizophrenia.J Psychiatry Neurosci. 2014 Sep;39(5):312-20. doi: 10.1503/jpn.130269. J Psychiatry Neurosci. 2014. PMID: 24866554 Free PMC article.
-
Dopamine 2 Receptor Genes Are Associated with Raised Blood Glucose in Schizophrenia.Can J Psychiatry. 2016 May;61(5):291-7. doi: 10.1177/0706743716644765. Epub 2016 Apr 6. Can J Psychiatry. 2016. PMID: 27254804 Free PMC article.
-
MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression, and the microRNA-mediated expression regulation is altered by a genetic variant.J Biol Chem. 2014 May 9;289(19):13434-44. doi: 10.1074/jbc.M113.535203. Epub 2014 Mar 27. J Biol Chem. 2014. PMID: 24675081 Free PMC article.
-
Allelic imbalance associated with the schizophrenia risk SNP rs1344706 indicates a cis-acting variant in ZNF804A.Schizophr Res. 2014 Mar;153(1-3):243-5. doi: 10.1016/j.schres.2014.01.005. Epub 2014 Jan 23. Schizophr Res. 2014. PMID: 24462263 Free PMC article. No abstract available.
References
-
- Berrettini W. Evidence for shared susceptibility in bipolar disorder and schizophrenia. Am. J. Med. Genet. C Semin. Med. Genet. 2003;123C:59–64. - PubMed
-
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q 11). Biochem. Mosc. 2007;72:578–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

