Genetic changes during a laboratory adaptive evolution process that allowed fast growth in glucose to an Escherichia coli strain lacking the major glucose transport system
- PMID: 22884033
- PMCID: PMC3469383
- DOI: 10.1186/1471-2164-13-385
Genetic changes during a laboratory adaptive evolution process that allowed fast growth in glucose to an Escherichia coli strain lacking the major glucose transport system
Abstract
Background: Escherichia coli strains lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS), which is the major bacterial component involved in glucose transport and its phosphorylation, accumulate high amounts of phosphoenolpyruvate that can be diverted to the synthesis of commercially relevant products. However, these strains grow slowly in glucose as sole carbon source due to its inefficient transport and metabolism. Strain PB12, with 400% increased growth rate, was isolated after a 120 hours adaptive laboratory evolution process for the selection of faster growing derivatives in glucose. Analysis of the genetic changes that occurred in the PB12 strain that lacks PTS will allow a better understanding of the basis of its growth adaptation and, therefore, in the design of improved metabolic engineering strategies for enhancing carbon diversion into the aromatic pathways.
Results: Whole genome analyses using two different sequencing methodologies: the Roche NimbleGen Inc. comparative genome sequencing technique, and high throughput sequencing with Illumina Inc. GAIIx, allowed the identification of the genetic changes that occurred in the PB12 strain. Both methods detected 23 non-synonymous and 22 synonymous point mutations. Several non-synonymous mutations mapped in regulatory genes (arcB, barA, rpoD, rna) and in other putative regulatory loci (yjjU, rssA and ypdA). In addition, a chromosomal deletion of 10,328 bp was detected that removed 12 genes, among them, the rppH, mutH and galR genes. Characterization of some of these mutated and deleted genes with their functions and possible functions, are presented.
Conclusions: The deletion of the contiguous rppH, mutH and galR genes that occurred simultaneously, is apparently the main reason for the faster growth of the evolved PB12 strain. In support of this interpretation is the fact that inactivation of the rppH gene in the parental PB11 strain substantially increased its growth rate, very likely by increasing glycolytic mRNA genes stability. Furthermore, galR inactivation allowed glucose transport by GalP into the cell. The deletion of mutH in an already stressed strain that lacks PTS is apparently responsible for the very high mutation rate observed.
Figures
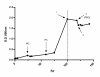

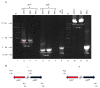

Similar articles
-
Deletion of the 2-acyl-glycerophosphoethanolamine cycle improve glucose metabolism in Escherichia coli strains employed for overproduction of aromatic compounds.Microb Cell Fact. 2015 Dec 1;14:194. doi: 10.1186/s12934-015-0382-6. Microb Cell Fact. 2015. PMID: 26627477 Free PMC article.
-
Analysis of differentially upregulated proteins in ptsHIcrr- and rppH- mutants in Escherichia coli during an adaptive laboratory evolution experiment.Appl Microbiol Biotechnol. 2018 Dec;102(23):10193-10208. doi: 10.1007/s00253-018-9397-3. Epub 2018 Oct 3. Appl Microbiol Biotechnol. 2018. PMID: 30284012
-
Adaptation for fast growth on glucose by differential expression of central carbon metabolism and gal regulon genes in an Escherichia coli strain lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system.Metab Eng. 2005 Mar;7(2):70-87. doi: 10.1016/j.ymben.2004.10.002. Metab Eng. 2005. PMID: 15781417
-
Inactivation of the PTS as a Strategy to Engineer the Production of Aromatic Metabolites in Escherichia coli.J Mol Microbiol Biotechnol. 2015;25(2-3):195-208. doi: 10.1159/000380854. Epub 2015 Jul 9. J Mol Microbiol Biotechnol. 2015. PMID: 26159079 Review.
-
Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation.Appl Microbiol Biotechnol. 2012 Jun;94(6):1483-94. doi: 10.1007/s00253-012-4101-5. Epub 2012 May 11. Appl Microbiol Biotechnol. 2012. PMID: 22573269 Review.
Cited by
-
Physiological and transcriptional characterization of Escherichia coli strains lacking interconversion of phosphoenolpyruvate and pyruvate when glucose and acetate are coutilized.Biotechnol Bioeng. 2014 Jun;111(6):1150-60. doi: 10.1002/bit.25177. Epub 2014 Jan 28. Biotechnol Bioeng. 2014. PMID: 24375081 Free PMC article.
-
An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies.BMC Genomics. 2014 Dec 13;15(1):1103. doi: 10.1186/1471-2164-15-1103. BMC Genomics. 2014. PMID: 25494611 Free PMC article.
-
A novel small RNA CoaR regulates coenzyme A biosynthesis and tolerance of Synechocystis sp. PCC6803 to 1-butanol possibly via promoter-directed transcriptional silencing.Biotechnol Biofuels. 2017 Feb 20;10:42. doi: 10.1186/s13068-017-0727-y. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28239414 Free PMC article.
-
A latent capacity for evolutionary innovation through exaptation in metabolic systems.Nature. 2013 Aug 8;500(7461):203-6. doi: 10.1038/nature12301. Epub 2013 Jul 14. Nature. 2013. PMID: 23851393
-
Enhanced Production of Pterostilbene in Escherichia coli Through Directed Evolution and Host Strain Engineering.Front Microbiol. 2021 Oct 7;12:710405. doi: 10.3389/fmicb.2021.710405. eCollection 2021. Front Microbiol. 2021. PMID: 34690954 Free PMC article.
References
-
- Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BØ. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

