A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern
- PMID: 22885008
- PMCID: PMC3472102
- DOI: 10.1016/j.molcel.2012.07.003
A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern
Abstract
Although the genomic pattern of nucleosome positioning is broadly conserved, quantitative aspects vary over evolutionary timescales. We identify the cis and trans determinants of nucleosome positioning using a functional evolutionary approach involving S. cerevisiae strains containing large genomic regions from other yeast species. In a foreign species, nucleosome depletion at promoters is maintained over poly(dA:dT) tracts, whereas internucleosome spacing and all other aspects of nucleosome positioning tested are not. Interestingly, the locations of the +1 nucleosome and RNA start sites shift in concert. Strikingly, in a foreign species, nucleosome-depleted regions occur fortuitously in coding regions, and they often act as promoters that are associated with a positioned nucleosome array linked to the length of the transcription unit. We suggest a three-step model in which nucleosome remodelers, general transcription factors, and the transcriptional elongation machinery are primarily involved in generating the nucleosome positioning pattern in vivo.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

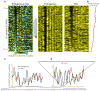

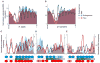


Comment in
-
Chromatin: a model for nucleosome positioning.Nat Rev Genet. 2012 Oct;13(10):674-5. doi: 10.1038/nrg3331. Epub 2012 Aug 29. Nat Rev Genet. 2012. PMID: 22929987 No abstract available.
-
Evolutionary insights into genome-wide nucleosome positioning.Genome Biol. 2012;13(9):170. doi: 10.1186/gb4046. Genome Biol. 2012. PMID: 23017016 Free PMC article.
-
Nucleosome positioning: multiple mechanisms toward a unifying goal.Mol Cell. 2012 Oct 12;48(1):1-2. doi: 10.1016/j.molcel.2012.09.015. Mol Cell. 2012. PMID: 23062951
Similar articles
-
Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters.Genome Res. 2010 Apr;20(4):473-84. doi: 10.1101/gr.103226.109. Epub 2010 Feb 4. Genome Res. 2010. PMID: 20133331 Free PMC article.
-
Nucleosome free regions in yeast promoters result from competitive binding of transcription factors that interact with chromatin modifiers.PLoS Comput Biol. 2013;9(8):e1003181. doi: 10.1371/journal.pcbi.1003181. Epub 2013 Aug 22. PLoS Comput Biol. 2013. PMID: 23990766 Free PMC article.
-
A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome.Science. 2011 May 20;332(6032):977-80. doi: 10.1126/science.1200508. Science. 2011. PMID: 21596991 Free PMC article.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo.Int J Mol Sci. 2021 Jul 30;22(15):8233. doi: 10.3390/ijms22158233. Int J Mol Sci. 2021. PMID: 34360997 Free PMC article. Review.
Cited by
-
Gene regulation by the act of long non-coding RNA transcription.BMC Biol. 2013 May 30;11:59. doi: 10.1186/1741-7007-11-59. BMC Biol. 2013. PMID: 23721193 Free PMC article. Review.
-
Dynamics of Nucleosome Positioning Maturation following Genomic Replication.Cell Rep. 2016 Sep 6;16(10):2651-2665. doi: 10.1016/j.celrep.2016.07.083. Epub 2016 Aug 25. Cell Rep. 2016. PMID: 27568571 Free PMC article.
-
Species-specific factors mediate extensive heterogeneity of mRNA 3' ends in yeasts.Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):11073-8. doi: 10.1073/pnas.1309384110. Epub 2013 Jun 17. Proc Natl Acad Sci U S A. 2013. PMID: 23776204 Free PMC article.
-
Establishment and Maintenance of Chromatin Architecture Are Promoted Independently of Transcription by the Histone Chaperone FACT and H3-K56 Acetylation in Saccharomyces cerevisiae.Genetics. 2019 Mar;211(3):877-892. doi: 10.1534/genetics.118.301853. Epub 2019 Jan 24. Genetics. 2019. PMID: 30679261 Free PMC article.
-
Nucleosome-positioning sequence repeats impact chromatin silencing in yeast minichromosomes.Genetics. 2014 Nov;198(3):1015-29. doi: 10.1534/genetics.114.169508. Epub 2014 Sep 3. Genetics. 2014. PMID: 25189873 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

