Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator
- PMID: 22885060
- PMCID: PMC3498763
- DOI: 10.1016/j.cub.2012.07.042
Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator
Abstract
Background: Ant societies comprise individuals belonging to different castes characterized by specialized morphologies and behaviors. Because ant embryos can follow different developmental trajectories, epigenetic mechanisms must play a role in caste determination. Ants have a full set of DNA methyltransferases and their genomes contain methylcytosine. To determine the relationship between DNA methylation and phenotypic plasticity in ants, we obtained and compared the genome-wide methylomes of different castes and developmental stages of Camponotus floridanus and Harpegnathos saltator.
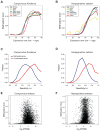
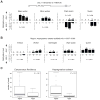
Results: In the ant genomes, methylcytosines are found both in symmetric CG dinucleotides (CpG) and non-CpG contexts and are strongly enriched at exons of active genes. Changes in exonic DNA methylation correlate with alternative splicing events such as exon skipping and alternative splice site selection. Several genes exhibit caste-specific and developmental changes in DNA methylation that are conserved between the two species, including genes involved in reproduction, telomere maintenance, and noncoding RNA metabolism. Several loci are methylated and expressed monoallelically, and in some cases, the choice of methylated allele depends on the caste.
Conclusions: These first ant methylomes and their intra- and interspecies comparison reveal an exonic methylation pattern that points to a connection between DNA methylation and splicing. The presence of monoallelic DNA methylation and the methylation of non-CpG sites in all samples suggest roles in genome regulation in these social insects, including the intriguing possibility of parental or caste-specific genomic imprinting.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures




Comment in
-
Epigenetics: the making of ant castes.Curr Biol. 2012 Oct 9;22(19):R835-8. doi: 10.1016/j.cub.2012.07.045. Curr Biol. 2012. PMID: 23058801
References
-
- Hölldobler B, Wilson EO. The Ants. Cambridge, MA: Harvard University Press; 1990.
-
- Bonasio R. Emerging topics in epigenetics: ants, brains, and noncoding RNAs. Ann N Y Acad Sci 2012 - PubMed
-
- Peeters C, Liebig J, Hölldobler B. Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insectes Sociaux. 2000;47:325–332.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

