Diversity and versatility of the Thermotoga maritima sugar kinome
- PMID: 22885293
- PMCID: PMC3458674
- DOI: 10.1128/JB.01136-12
Diversity and versatility of the Thermotoga maritima sugar kinome
Abstract
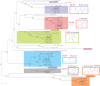
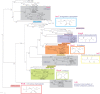
Sugar phosphorylation is an indispensable committed step in a large variety of sugar catabolic pathways, which are major suppliers of carbon and energy in heterotrophic species. Specialized sugar kinases that are indispensable for most of these pathways can be utilized as signature enzymes for the reconstruction of carbohydrate utilization machinery from microbial genomic and metagenomic data. Sugar kinases occur in several structurally distinct families with various partially overlapping as well as yet unknown substrate specificities that often cannot be accurately assigned by homology-based techniques. A subsystems-based metabolic reconstruction combined with the analysis of genome context and followed by experimental testing of predicted gene functions is a powerful approach of functional gene annotation. Here we applied this integrated approach for functional mapping of all sugar kinases constituting an extensive and diverse sugar kinome in the thermophilic bacterium Thermotoga maritima. Substrate preferences of 14 kinases mainly from the FGGY and PfkB families were inferred by bioinformatics analysis and biochemically characterized by screening with a panel of 45 different carbohydrates. Most of the analyzed enzymes displayed narrow substrate preferences corresponding to their predicted physiological roles in their respective catabolic pathways. The observed consistency supports the choice of kinases as signature enzymes for genomics-based identification and reconstruction of sugar utilization pathways. Use of the integrated genomic and experimental approach greatly speeds up the identification of the biochemical function of unknown proteins and improves the quality of reconstructed pathways.
Figures

References
-
- Andreassi JL, II, Leyh TS. 2004. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry 43:14594–14601 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

