H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes
- PMID: 22885324
- PMCID: PMC4533924
- DOI: 10.1038/nsmb.2356
H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes
Abstract
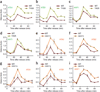
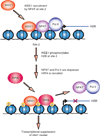
Histone gene transcription is actively downregulated after completion of DNA synthesis to avoid overproduction. However, the precise mechanistic details of the cessation of histone mRNA synthesis are not clear. We found that histone H2B phosphorylation at Tyr37 occurs upstream of histone cluster 1, Hist1, during the late S phase. We identified WEE1 as the kinase that phosphorylates H2B at Tyr37. Loss of expression or inhibition of WEE1 kinase abrogated H2B Tyr37 phosphorylation with a concomitant increase in histone transcription in yeast and mammalian cells. H2B Tyr37 phosphorylation excluded binding of the transcriptional coactivator NPAT and RNA polymerase II and recruited the histone chaperone HIRA upstream of the Hist1 cluster. Taken together, our data show a previously unknown and evolutionarily conserved function for WEE1 kinase as an epigenetic modulator that marks chromatin with H2B Tyr37 phosphorylation, thereby inhibiting the transcription of multiple histone genes to lower the burden on the histone mRNA turnover machinery.
Figures



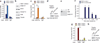
Similar articles
-
WEE1 tyrosine kinase, a novel epigenetic modifier.Trends Genet. 2013 Jul;29(7):394-402. doi: 10.1016/j.tig.2013.02.003. Epub 2013 Mar 26. Trends Genet. 2013. PMID: 23537585 Free PMC article. Review.
-
Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation.J Mol Biol. 2014 Aug 12;426(16):2928-2941. doi: 10.1016/j.jmb.2014.05.028. Epub 2014 Jun 6. J Mol Biol. 2014. PMID: 24911582 Free PMC article.
-
Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein.Genes Dev. 2011 Dec 1;25(23):2489-501. doi: 10.1101/gad.173427.111. Genes Dev. 2011. PMID: 22156209 Free PMC article.
-
Histone H2B ubiquitylation is associated with elongating RNA polymerase II.Mol Cell Biol. 2005 Jan;25(2):637-51. doi: 10.1128/MCB.25.2.637-651.2005. Mol Cell Biol. 2005. PMID: 15632065 Free PMC article.
-
Regulation of histone gene transcription in yeast.Cell Mol Life Sci. 2014 Feb;71(4):599-613. doi: 10.1007/s00018-013-1443-9. Epub 2013 Aug 23. Cell Mol Life Sci. 2014. PMID: 23974242 Free PMC article. Review.
Cited by
-
Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.Nat Commun. 2022 Nov 14;13(1):6929. doi: 10.1038/s41467-022-34724-5. Nat Commun. 2022. PMID: 36376335 Free PMC article.
-
Combined Aurora Kinase A (AURKA) and WEE1 Inhibition Demonstrates Synergistic Antitumor Effect in Squamous Cell Carcinoma of the Head and Neck.Clin Cancer Res. 2019 Jun 1;25(11):3430-3442. doi: 10.1158/1078-0432.CCR-18-0440. Epub 2019 Feb 12. Clin Cancer Res. 2019. PMID: 30755439 Free PMC article.
-
Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation.Genome Res. 2013 Oct;23(10):1563-79. doi: 10.1101/gr.154872.113. Epub 2013 Jul 26. Genome Res. 2013. PMID: 23893515 Free PMC article.
-
Epigenetics of inflammation, maternal infection, and nutrition.J Nutr. 2015 May;145(5):1109S-1115S. doi: 10.3945/jn.114.194639. Epub 2015 Apr 1. J Nutr. 2015. PMID: 25833887 Free PMC article. Review.
-
Functional Impact of the H2A.Z Histone Variant During Meiosis in Saccharomyces cerevisiae.Genetics. 2018 Aug;209(4):997-1015. doi: 10.1534/genetics.118.301110. Epub 2018 May 31. Genetics. 2018. PMID: 29853474 Free PMC article.
References
-
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. - PubMed
-
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. - PubMed
-
- Borun TW, Gabrielli F, Ajiro K, Zweidler A, Baglioni C. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975;4:59–67. - PubMed
-
- Hereford L, Bromley S, Osley MA. Periodic transcription of yeast histone genes. Cell. 1982;30:305–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

