HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle
- PMID: 22885700
- PMCID: PMC3443318
- DOI: 10.1038/nature11316
HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle
Abstract
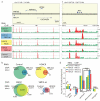
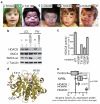
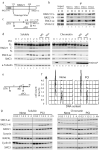
Cornelia de Lange syndrome (CdLS) is a dominantly inherited congenital malformation disorder, caused by mutations in the cohesin-loading protein NIPBL for nearly 60% of individuals with classical CdLS, and by mutations in the core cohesin components SMC1A (~5%) and SMC3 (<1%) for a smaller fraction of probands. In humans, the multisubunit complex cohesin is made up of SMC1, SMC3, RAD21 and a STAG protein. These form a ring structure that is proposed to encircle sister chromatids to mediate sister chromatid cohesion and also has key roles in gene regulation. SMC3 is acetylated during S-phase to establish cohesiveness of chromatin-loaded cohesin, and in yeast, the class I histone deacetylase Hos1 deacetylates SMC3 during anaphase. Here we identify HDAC8 as the vertebrate SMC3 deacetylase, as well as loss-of-function HDAC8 mutations in six CdLS probands. Loss of HDAC8 activity results in increased SMC3 acetylation and inefficient dissolution of the ‘used’ cohesin complex released from chromatin in both prophase and anaphase. SMC3 with retained acetylation is loaded onto chromatin, and chromatin immunoprecipitation sequencing analysis demonstrates decreased occupancy of cohesin localization sites that results in a consistent pattern of altered transcription seen in CdLS cell lines with either NIPBL or HDAC8 mutations.
Figures

References
-
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–603. - PubMed
-
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–6. - PubMed
-
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–41. - PubMed
Additional Methods References
-
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–3. - PubMed
-
- Watrin E, et al. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–74. - PubMed
-
- Strom T. ExonPrimer. 2006
-
- Eddy SR. Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol. 1995;3:114–20. - PubMed
-
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

