Proteogenomic elucidation of the initial steps in the benzene degradation pathway of a novel halophile, Arhodomonas sp. strain Rozel, isolated from a hypersaline environment
- PMID: 22885747
- PMCID: PMC3457091
- DOI: 10.1128/AEM.01327-12
Proteogenomic elucidation of the initial steps in the benzene degradation pathway of a novel halophile, Arhodomonas sp. strain Rozel, isolated from a hypersaline environment
Abstract
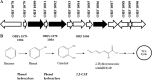
Lately, there has been a special interest in understanding the role of halophilic and halotolerant organisms for their ability to degrade hydrocarbons. The focus of this study was to investigate the genes and enzymes involved in the initial steps of the benzene degradation pathway in halophiles. The extremely halophilic bacteria Arhodomonas sp. strain Seminole and Arhodomonas sp. strain Rozel, which degrade benzene and toluene as the sole carbon source at high salinity (0.5 to 4 M NaCl), were isolated from enrichments developed from contaminated hypersaline environments. To obtain insights into the physiology of this novel group of organisms, a draft genome sequence of the Seminole strain was obtained. A cluster of 13 genes predicted to be functional in the hydrocarbon degradation pathway was identified from the sequence. Two-dimensional (2D) gel electrophoresis and liquid chromatography-mass spectrometry were used to corroborate the role of the predicted open reading frames (ORFs). ORFs 1080 and 1082 were identified as components of a multicomponent phenol hydroxylase complex, and ORF 1086 was identified as catechol 2,3-dioxygenase (2,3-CAT). Based on this analysis, it was hypothesized that benzene is converted to phenol and then to catechol by phenol hydroxylase components. The resulting catechol undergoes ring cleavage via the meta pathway by 2,3-CAT to form 2-hydroxymuconic semialdehyde, which enters the tricarboxylic acid cycle. To substantiate these findings, the Rozel strain was grown on deuterated benzene, and gas chromatography-mass spectrometry detected deuterated phenol as the initial intermediate of benzene degradation. These studies establish the initial steps of the benzene degradation pathway in halophiles.
Figures



Similar articles
-
Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments.Front Microbiol. 2014 Apr 23;5:173. doi: 10.3389/fmicb.2014.00173. eCollection 2014. Front Microbiol. 2014. PMID: 24795705 Free PMC article. Review.
-
Arhodomonas sp. strain Seminole and its genetic potential to degrade aromatic compounds under high-salinity conditions.Appl Environ Microbiol. 2014 Nov;80(21):6664-76. doi: 10.1128/AEM.01509-14. Epub 2014 Aug 22. Appl Environ Microbiol. 2014. PMID: 25149520 Free PMC article.
-
Biodegradation of BTEX at high salinity by an enrichment culture from hypersaline sediments of Rozel Point at Great Salt Lake.J Appl Microbiol. 2009 Dec 1;107(6):2001-8. doi: 10.1111/j.1365-2672.2009.04385.x. Epub 2009 May 20. J Appl Microbiol. 2009. PMID: 19519667
-
The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol.Appl Environ Microbiol. 1999 Apr;65(4):1589-95. doi: 10.1128/AEM.65.4.1589-1595.1999. Appl Environ Microbiol. 1999. PMID: 10103255 Free PMC article.
-
Biodegradation of petroleum hydrocarbons in hypersaline environments.Braz J Microbiol. 2012 Jul;43(3):865-72. doi: 10.1590/S1517-83822012000300003. Epub 2012 Jun 1. Braz J Microbiol. 2012. PMID: 24031900 Free PMC article. Review.
Cited by
-
Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments.Front Microbiol. 2014 Apr 23;5:173. doi: 10.3389/fmicb.2014.00173. eCollection 2014. Front Microbiol. 2014. PMID: 24795705 Free PMC article. Review.
-
Evidence for surfactant production by the haloarchaeon Haloferax sp. MSNC14 in hydrocarbon-containing media.Extremophiles. 2013 Jul;17(4):669-75. doi: 10.1007/s00792-013-0550-8. Epub 2013 Jun 9. Extremophiles. 2013. PMID: 23748377
-
Metagenomic analysis reveals specific BTEX degrading microorganisms of a bacterial consortium.AMB Express. 2023 May 17;13(1):48. doi: 10.1186/s13568-023-01541-y. AMB Express. 2023. PMID: 37195357 Free PMC article.
-
Isolation and characterization of two novel halotolerant Catechol 2, 3-dioxygenases from a halophilic bacterial consortium.Sci Rep. 2015 Dec 1;5:17603. doi: 10.1038/srep17603. Sci Rep. 2015. PMID: 26621792 Free PMC article.
-
Arhodomonas sp. strain Seminole and its genetic potential to degrade aromatic compounds under high-salinity conditions.Appl Environ Microbiol. 2014 Nov;80(21):6664-76. doi: 10.1128/AEM.01509-14. Epub 2014 Aug 22. Appl Environ Microbiol. 2014. PMID: 25149520 Free PMC article.
References
-
- Aktas DF, et al. 2010. Anaerobic metabolism of biodiesel and its impact on metal corrosion. Energy Fuels 24:2924–2928
-
- Al-Mailem DM, Sorkhoh NA, Al-Awadhi H, Eliyas M, Radwan SS. 2010. Biodegradation of crude oil and pure hydrocarbons by extreme halophilic archaea from hypersaline coasts of the Arabian Gulf. Extremophiles 14:321–328 - PubMed
-
- Al-Mueini R, Al-Dalali M, Al-Amri IS, Patzelt H. 2007. Hydrocarbon degradation at high salinity by a novel extremely halophilic actinomycete. Environ. Chem. 4:5–7
-
- Bastos AE, Moon DH, Rossi A, Trevors JT, Tsai SM. 2000. Salt-tolerant phenol-degrading microorganisms isolated from Amazonian soil samples. Arch. Microbiol. 174:346–352 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

