A metabolic prosurvival role for PML in breast cancer
- PMID: 22886304
- PMCID: PMC3433768
- DOI: 10.1172/JCI62129
A metabolic prosurvival role for PML in breast cancer
Abstract
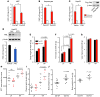
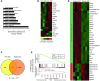
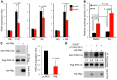
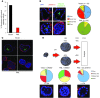
Cancer cells exhibit an aberrant metabolism that facilitates more efficient production of biomass and hence tumor growth and progression. However, the genetic cues modulating this metabolic switch remain largely undetermined. We identified a metabolic function for the promyelocytic leukemia (PML) gene, uncovering an unexpected role for this bona fide tumor suppressor in breast cancer cell survival. We found that PML acted as both a negative regulator of PPARγ coactivator 1A (PGC1A) acetylation and a potent activator of PPAR signaling and fatty acid oxidation. We further showed that PML promoted ATP production and inhibited anoikis. Importantly, PML expression allowed luminal filling in 3D basement membrane breast culture models, an effect that was reverted by the pharmacological inhibition of fatty acid oxidation. Additionally, immunohistochemical analysis of breast cancer biopsies revealed that PML was overexpressed in a subset of breast cancers and enriched in triple-negative cases. Indeed, PML expression in breast cancer correlated strikingly with reduced time to recurrence, a gene signature of poor prognosis, and activated PPAR signaling. These findings have important therapeutic implications, as PML and its key role in fatty acid oxidation metabolism are amenable to pharmacological suppression, a potential future mode of cancer prevention and treatment.
Figures



References
-
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Cancer. 2011;11(5):325–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

