ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets
- PMID: 22887839
- PMCID: PMC3448863
- DOI: 10.1002/stem.1193
ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets
Abstract
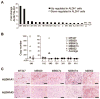
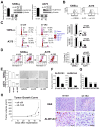
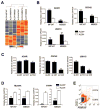
Although the concept of cancer stem cells (CSCs) is well-accepted for many tumors, the existence of such cells in human melanoma has been the subject of debate. In this study, we demonstrate the existence of human melanoma cells that fulfill the criteria for CSCs (self-renewal and differentiation) by serially xenotransplanting cells into nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice. These cells possess high aldehyde dehydrogenase (ALDH) activity with ALDH1A1 and ALDH1A3 being the predominant ALDH isozymes. ALDH-positive melanoma cells are more tumorigenic than ALDH-negative cells in both NOD/SCID mice and NSG mice. Biological analyses of the ALDH-positive melanoma cells reveal the ALDH isozymes to be key molecules regulating the function of these cells. Silencing ALDH1A by siRNA or shRNA leads to cell cycle arrest, apoptosis, decreased cell viability in vitro, and reduced tumorigenesis in vivo. ALDH-positive melanoma cells are more resistant to chemotherapeutic agents and silencing ALDH1A by siRNA sensitizes melanoma cells to drug-induced cell death. Furthermore, we, for the first time, examined the molecular signatures of ALDH-positive CSCs from patient-derived tumor specimens. The signatures of melanoma CSCs include retinoic acid (RA)-driven target genes with RA response elements and genes associated with stem cell function. These findings implicate that ALDH isozymes are not only biomarkers of CSCs but also attractive therapeutic targets for human melanoma. Further investigation of these isozymes and genes will enhance our understanding of the molecular mechanisms governing CSCs and reveal new molecular targets for therapeutic intervention of cancer.
Copyright © 2012 AlphaMed Press.
Conflict of interest statement
The authors disclosed no potential conflicts of interest.
Figures


References
-
- Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. - PubMed
-
- Fang D, Nguyen TK, Leishear K, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. - PubMed
-
- Monzani E, Facchetti F, Galmozzi E, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

