The apo-structure of the low molecular weight protein-tyrosine phosphatase A (MptpA) from Mycobacterium tuberculosis allows for better target-specific drug development
- PMID: 22888002
- PMCID: PMC3464563
- DOI: 10.1074/jbc.M112.399261
The apo-structure of the low molecular weight protein-tyrosine phosphatase A (MptpA) from Mycobacterium tuberculosis allows for better target-specific drug development
Abstract
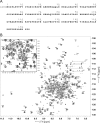
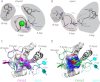
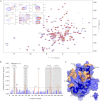
Protein-tyrosine phosphatases (PTPs) and protein-tyrosine kinases co-regulate cellular processes. In pathogenic bacteria, they are frequently exploited to act as key virulence factors for human diseases. Mycobacterium tuberculosis, the causative organism of tuberculosis, secretes a low molecular weight PTP (LMW-PTP), MptpA, which is required for its survival upon infection of host macrophages. Although there is otherwise no sequence similarity of LMW-PTPs to other classes of PTPs, the phosphate binding loop (P-loop) CX(5)R and the loop containing a critical aspartic acid residue (D-loop), required for the catalytic activity, are well conserved. In most high molecular weight PTPs, ligand binding to the P-loop triggers a large conformational reorientation of the D-loop, in which it moves ∼10 Å, from an "open" to a "closed" conformation. Until now, there have been no ligand-free structures of LMW-PTPs described, and hence the dynamics of the D-loop have remained largely unknown for these PTPs. Here, we present a high resolution solution NMR structure of the free form of the MptpA LMW-PTP. In the absence of ligand and phosphate ions, the D-loop adopts an open conformation. Furthermore, we characterized the binding site of phosphate, a competitive inhibitor of LMW-PTPs, on MptpA and elucidated the involvement of both the P- and D-loop in phosphate binding. Notably, in LMW-PTPs, the phosphorylation status of two well conserved tyrosine residues, typically located in the D-loop, regulates the enzyme activity. PtkA, the kinase complementary to MptpA, phosphorylates these two tyrosine residues in MptpA. We characterized the MptpA-PtkA interaction by NMR spectroscopy to show that both the P- and D-loop form part of the binding interface.
Figures




References
-
- World Health Organization (2012) 2011/2012 Tuberculosis Global Facts, World Health Organization, Geneva
-
- De Cock K. M., Chaisson R. E. (1999) Will DOTS do it? A reappraisal of tuberculosis control in countries with high rates of HIV infection. Int. J. Tuberc. Lung Dis. 3, 457–465 - PubMed
-
- Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. (2011) The challenge of new drug discovery for tuberculosis. Nature 469, 483–490 - PubMed
-
- Clatworthy A. E., Pierson E., Hung D. T. (2007) Targeting virulence. A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 - PubMed
-
- Cozzone A. J., Grangeasse C., Doublet P., Duclos B. (2004) Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181, 171–181 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

