Inhibition of Yersinia pestis DNA adenine methyltransferase in vitro by a stibonic acid compound: identification of a potential novel class of antimicrobial agents
- PMID: 22889062
- PMCID: PMC3570013
- DOI: 10.1111/j.1476-5381.2012.02134.x
Inhibition of Yersinia pestis DNA adenine methyltransferase in vitro by a stibonic acid compound: identification of a potential novel class of antimicrobial agents
Abstract
Background and purpose: Multiple antibiotic resistant strains of plague are emerging, driving a need for the development of novel antibiotics effective against Yersinia pestis. DNA adenine methylation regulates numerous fundamental processes in bacteria and alteration of DNA adenine methlytransferase (Dam) expression is attenuating for several pathogens, including Y. pestis. The lack of a functionally similar enzyme in humans makes Dam a suitable target for development of novel therapeutics for plague.
Experimental approach: Compounds were evaluated for their ability to inhibit Dam activity in a high-throughput screening assay. DNA was isolated from Yersinia grown in the presence of lead compounds and restricted to determine the effect of inhibitors on DNA methylation. Transcriptional analysis was undertaken to determine the effect of an active inhibitor on virulence-associated phenotypes.
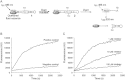
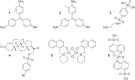
Key results: We have identified a series of aryl stibonic acids which inhibit Dam in vitro. The most active, 4-stibonobenzenesulfonic acid, exhibited a competitive mode of inhibition with respect to DNA and a K(i) of 6.46 nM. One compound was found to inhibit DNA methylation in cultured Y. pestis. The effects of this inhibition on the physiology of the cell were widespread, and included altered expression of known virulence traits, including iron acquisition and Type III secretion.
Conclusions and implications: We have identified a novel class of potent Dam inhibitors. Treatment of bacterial cell cultures with these inhibitors resulted in a decrease in DNA methylation. Expression of virulence factors was affected, suggesting these inhibitors may attenuate bacterial infectivity and function as antibiotics.
© 2012 The Authors. British Journal of Pharmacology © 2012 The British Pharmacological Society.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

