Probing the role of stochasticity in a model of the embryonic stem cell: heterogeneous gene expression and reprogramming efficiency
- PMID: 22889237
- PMCID: PMC3468383
- DOI: 10.1186/1752-0509-6-98
Probing the role of stochasticity in a model of the embryonic stem cell: heterogeneous gene expression and reprogramming efficiency
Abstract
Background: Embryonic stem cells (ESC) have the capacity to self-renew and remain pluripotent, while continuously providing a source of a variety of differentiated cell types. Understanding what governs these properties at the molecular level is crucial for stem cell biology and its application to regenerative medicine. Of particular relevance is to elucidate those molecular interactions which govern the reprogramming of somatic cells into ESC. A computational approach can be used as a framework to explore the dynamics of a simplified network of the ESC with the aim to understand how stem cells differentiate and also how they can be reprogrammed from somatic cells.
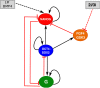
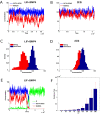
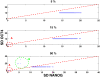
Results: We propose a computational model of the embryonic stem cell network, in which a core set of transcription factors (TFs) interact with each other and are induced by external factors. A stochastic treatment of the network dynamics suggests that NANOG heterogeneity is the deciding factor for the stem cell fate. In particular, our results show that the decision of staying in the ground state or commitment to a differentiated state is fundamentally stochastic, and can be modulated by the addition of external factors (2i/3i media), which have the effect of reducing fluctuations in NANOG expression. Our model also hosts reprogramming of a committed cell into an ESC by over-expressing OCT4. In this context, we recapitulate the important experimental result that reprogramming efficiency peaks when OCT4 is over-expressed within a specific range of values.
Conclusions: We have demonstrated how a stochastic computational model based upon a simplified network of TFs in ESCs can elucidate several key observed dynamical features. It accounts for (i) the observed heterogeneity of key regulators, (ii) characterizes the ESC under certain external stimuli conditions and (iii) describes the occurrence of transitions from the ESC to the differentiated state. Furthermore, the model (iv) provides a framework for reprogramming from somatic cells and conveys an understanding of reprogramming efficiency as a function of OCT4 over-expression.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

