An evolutionary perspective on the kinome of malaria parasites
- PMID: 22889911
- PMCID: PMC3415840
- DOI: 10.1098/rstb.2012.0014
An evolutionary perspective on the kinome of malaria parasites
Abstract
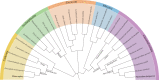
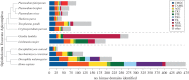
Malaria parasites belong to an ancient lineage that diverged very early from the main branch of eukaryotes. The approximately 90-member plasmodial kinome includes a majority of eukaryotic protein kinases that clearly cluster within the AGC, CMGC, TKL, CaMK and CK1 groups found in yeast, plants and mammals, testifying to the ancient ancestry of these families. However, several hundred millions years of independent evolution, and the specific pressures brought about by first a photosynthetic and then a parasitic lifestyle, led to the emergence of unique features in the plasmodial kinome. These include taxon-restricted kinase families, and unique peculiarities of individual enzymes even when they have homologues in other eukaryotes. Here, we merge essential aspects of all three malaria-related communications that were presented at the Evolution of Protein Phosphorylation meeting, and propose an integrated discussion of the specific features of the parasite's kinome and phosphoproteome.
Figures
References
-
- Kwiatkowski D. P. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria? Am. J. Hum. Genet. 77, 171–192 10.1086/432519 (doi:10.1086/432519) - DOI - DOI - PMC - PubMed
-
- Winstanley P., Ward S. 2006. Malaria chemotherapy. Adv. Parasitol. 61, 47–76 10.1016/S0065-308X(05)61002-0 (doi:10.1016/S0065-308X(05)61002-0) - DOI - DOI - PubMed
-
- Wells T. N., Poll E. M. 2010. When is enough enough? The need for a robust pipeline of high-quality antimalarials. Discov. Med. 9, 389–398 - PubMed
-
- Burrows J. N., Chibale K., Wells T. N. 2011. The state of the art in anti-malarial drug discovery and development. Curr. Top. Med. Chem. 11, 1226–1254 10.2174/156802611795429194 (doi:10.2174/156802611795429194) - DOI - DOI - PubMed
-
- Dondorp A. M., Yeung S., White L., Nguon C., Day N. P., Socheat D., von Seidlein L. 2010. Artemisinin resistance: current status and scenarios for containment. Nat. Rev. Microbiol. 8, 272–280 10.1038/nrmicro2385 (doi:10.1038/nrmicro2385) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

