From yeast to mammals: recent advances in genetic control of homologous recombination
- PMID: 22889934
- PMCID: PMC3468695
- DOI: 10.1016/j.dnarep.2012.07.001
From yeast to mammals: recent advances in genetic control of homologous recombination
Abstract
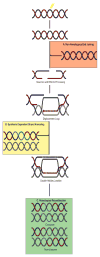
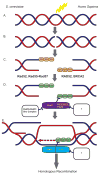
Misregulation of DNA repair is associated with genetic instability and tumorigenesis. To preserve the integrity of the genome, eukaryotic cells have evolved extremely intricate mechanisms for repairing DNA damage. One type of DNA lesion is a double-strand break (DSB), which is highly toxic when unrepaired. Repair of DSBs can occur through multiple mechanisms. Aside from religating the DNA ends, a homologous template can be used for repair in a process called homologous recombination (HR). One key step in committing to HR is the formation of Rad51 filaments, which perform the homology search and strand invasion steps. In S. cerevisiae, Srs2 is a key regulator of Rad51 filament formation and disassembly. In this review, we highlight potential candidates of Srs2 orthologues in human cells, and we discuss recent advances in understanding how Srs2's so-called "anti-recombinase" activity is regulated.
Copyright © 2012 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors confirm there is no conflict of interest, financial or otherwise, in this work.
Figures

References
-
- Duker NJ. Chromosome breakage syndromes and cancer. Am J Med Genet. 2002;115:125–129. - PubMed
-
- Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 2010;9:1256–1263. - PubMed
-
- Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 2010;9:1264–1272. - PubMed
-
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

