On rate limitations of electron transfer in the photosynthetic cytochrome b6f complex
- PMID: 22890107
- PMCID: PMC3505453
- DOI: 10.1039/c2cp41386h
On rate limitations of electron transfer in the photosynthetic cytochrome b6f complex
Abstract
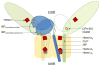
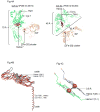
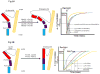
Considering information in the crystal structures of the cytochrome b(6)f complex relevant to the rate-limiting step in oxygenic photosynthesis, it is enigmatic that electron transport in the complex is not limited by the large distance, approximately 26 Å, between the iron-sulfur cluster (ISP) and its electron acceptor, cytochrome f. This enigma has been explained for the respiratory bc(1) complex by a crystal structure with a greatly shortened cluster-heme c(1) distance, leading to a concept of ISP dynamics in which the ISP soluble domain undergoes a translation-rotation conformation change and oscillates between positions relatively close to the cyt c(1) heme and a membrane-proximal position close to the ubiquinol electron-proton donor. Comparison of cytochrome b(6)f structures shows a variation in cytochrome f heme position that suggests the possibility of flexibility and motion of the extended cytochrome f structure that could entail a transient decrease in cluster-heme f distance. The dependence of cyt f turnover on lumen viscosity is consistent with a role of ISP - cyt f dynamics in determination of the rate-limiting step under conditions of low light intensity. Under conditions of low light intensity and proton electrochemical gradient present, for example, under a leaf canopy, it is proposed that a rate limitation of electron transport in the b(6)f complex may also arise from steric constraints in the entry/exit portal for passage of the plastoquinol and -quinone to/from its oxidation site proximal to the iron-sulfur cluster.
Figures
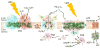



References
-
- Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science (New York, NY. 2003;302:1009–1014. - PubMed
-
- Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical heam in the cytochrome b6f complex. Nature. 2003;426:413–418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

