Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae
- PMID: 22890996
- PMCID: PMC3486043
- DOI: 10.1128/IAI.00647-12
Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae
Abstract
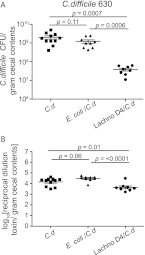
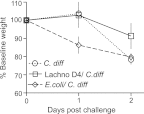
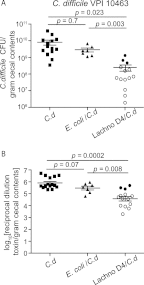
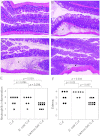
The indigenous microbial community of the gastrointestinal (GI) tract determines susceptibility to Clostridium difficile colonization and disease. Previous studies have demonstrated that antibiotic-treated mice challenged with C. difficile either developed rapidly lethal C. difficile infection or were stably colonized with mild disease. The GI microbial community of animals with mild disease was dominated by members of the bacterial family Lachnospiraceae, while the gut community in moribund animals had a predominance of Escherichia coli. We investigated the roles of murine Lachnospiraceae and E. coli strains in colonization resistance against C. difficile in germfree mice. Murine Lachnospiraceae and E. coli isolates were cultured from wild-type mice. The ability of each of these isolates to interfere with C. difficile colonization was tested by precolonizing germfree mice with these bacteria 4 days prior to experimental C. difficile challenge. Mice precolonized with a murine Lachnospiraceae isolate, but not those colonized with E. coli, had significantly decreased C. difficile colonization, lower intestinal cytotoxin levels and exhibited less severe clinical signs and colonic histopathology. Infection of germfree mice or mice precolonized with E. coli with C. difficile strain VPI 10463 was uniformly fatal by 48 h, but only 20% mortality was seen at 2 days in mice precolonized with the Lachnospiraceae isolate prior to challenge with VPI 10463. These findings confirm that a single component of the GI microbiota, a murine Lachnospiraceae isolate, could partially restore colonization resistance against C. difficile. Further study of the members within the Lachnospiraceae family could lead to a better understanding of mechanisms of colonization resistance against C. difficile and novel therapeutic approaches for the treatment and prevention of C. difficile infection.
Figures


References
-
- Bartlett JG. 2010. Clostridium difficile: progress and challenges. Ann. N. Y. Acad. Sci. 1213:62–69 - PubMed
-
- Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J. Infect. Dis. 136:701–705 - PubMed
-
- Chen X, et al. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

