Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides
- PMID: 22891237
- PMCID: PMC3461546
- DOI: 10.1104/pp.112.202309
Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides
Abstract
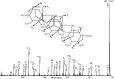
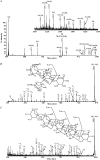
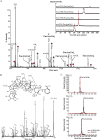
Proteins decorated with arabinogalactan (AG) have important roles in cell wall structure and plant development, yet the structure and biosynthesis of this polysaccharide are poorly understood. To facilitate the analysis of biosynthetic mutants, water-extractable arabinogalactan proteins (AGPs) were isolated from the leaves of Arabidopsis (Arabidopsis thaliana) plants and the structure of the AG carbohydrate component was studied. Enzymes able to hydrolyze specifically AG were utilized to release AG oligosaccharides. The released oligosaccharides were characterized by high-energy matrix-assisted laser desorption ionization-collision-induced dissociation mass spectrometry and polysaccharide analysis by carbohydrate gel electrophoresis. The Arabidopsis AG is composed of a β-(1→3)-galactan backbone with β-(1→6)-d-galactan side chains. The β-(1→6)-galactan side chains vary in length from one to over 20 galactosyl residues, and they are partly substituted with single α-(1→3)-l-arabinofuranosyl residues. Additionally, a substantial proportion of the β-(1→6)-galactan side chain oligosaccharides are substituted at the nonreducing termini with single 4-O-methyl-glucuronosyl residues via β-(1→6)-linkages. The β-(1→6)-galactan side chains are occasionally substituted with α-l-fucosyl. In the fucose-deficient murus1 mutant, AGPs lack these fucose modifications. This work demonstrates that Arabidopsis mutants in AGP structure can be identified and characterized. The detailed structural elucidation of the AG polysaccharides from the leaves of Arabidopsis is essential for insights into the structure-function relationships of these molecules and will assist studies on their biosynthesis.
Figures




References
-
- Chai W, Piskarev V, Lawson AM. (2001) Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal Chem 73: 651–657 - PubMed
-
- Cheung AY, Wang H, Wu HM. (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393 - PubMed
-
- Chun H, Shin DH, Hong BS, Cho HY, Yang HC. (2001) Purification and biological activity of acidic polysaccharide from leaves of Thymus vulgaris L. Biol Pharm Bull 24: 941–946 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

