Structure and assembly of a paramyxovirus matrix protein
- PMID: 22891297
- PMCID: PMC3435216
- DOI: 10.1073/pnas.1210275109
Structure and assembly of a paramyxovirus matrix protein
Abstract
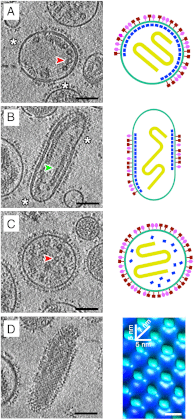
Many pleomorphic, lipid-enveloped viruses encode matrix proteins that direct their assembly and budding, but the mechanism of this process is unclear. We have combined X-ray crystallography and cryoelectron tomography to show that the matrix protein of Newcastle disease virus, a paramyxovirus and relative of measles virus, forms dimers that assemble into pseudotetrameric arrays that generate the membrane curvature necessary for virus budding. We show that the glycoproteins are anchored in the gaps between the matrix proteins and that the helical nucleocapsids are associated in register with the matrix arrays. About 90% of virions lack matrix arrays, suggesting that, in agreement with previous biological observations, the matrix protein needs to dissociate from the viral membrane during maturation, as is required for fusion and release of the nucleocapsid into the host's cytoplasm. Structure and sequence conservation imply that other paramyxovirus matrix proteins function similarly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1449–1496.
-
- McGinnes LW, Pantua H, Reitter J, Morrison TG. Newcastle disease virus: Propagation, quantification, and storage. Curr Protoc Microbiol. 2006 Chapter 15:Unit 15F.2. - PubMed
-
- National Select Agent Registry. http://www.selectagents.gov. Accessed August 2012.
-
- Scheid A, Caliguiri LA, Compans RW, Choppin PW. Isolation of paramyxovirus glycoproteins. Association of both hemagglutinating and neuraminidase activities with the larger SV5 glycoprotein. Virology. 1972;50:640–652. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

