Identification of a cis-acting DNA-protein interaction implicated in singular var gene choice in Plasmodium falciparum
- PMID: 22891919
- PMCID: PMC3549481
- DOI: 10.1111/cmi.12004
Identification of a cis-acting DNA-protein interaction implicated in singular var gene choice in Plasmodium falciparum
Abstract
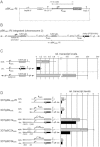
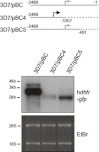
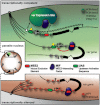
Plasmodium falciparum is responsible for the most severe form of malaria in humans. Antigenic variation of P. falciparum erythrocyte membrane protein 1 leads to immune evasion and occurs through switches in mutually exclusive var gene transcription. The recent progress in Plasmodium epigenetics notwithstanding, the mechanisms by which singularity of var activation is achieved are unknown. Here, we employed a functional approach to dissect the role of var gene upstream regions in mutually exclusive activation. Besides identifying sequence elements involved in activation and initiation of transcription, we mapped a region downstream of the transcriptional start site that is required to maintain singular var gene choice. Activation of promoters lacking this sequence occurs no longer in competition with endogenous var genes. Within this region we pinpointed a sequence-specific DNA-protein interaction involving a cis-acting sequence motif that is conserved in the majority of var loci. These results suggest an important role for this interaction in mutually exclusive locus recognition. Our findings are furthermore consistent with a novel mechanism for the control of singular gene choice in eukaryotes. In addition to their importance in P. falciparum antigenic variation, our results may also help to explain similar processes in other systems.
© 2012 Blackwell Publishing Ltd.
Figures



Similar articles
-
Insulator-like pairing elements regulate silencing and mutually exclusive expression in the malaria parasite Plasmodium falciparum.Proc Natl Acad Sci U S A. 2012 Dec 26;109(52):E3678-86. doi: 10.1073/pnas.1214572109. Epub 2012 Nov 29. Proc Natl Acad Sci U S A. 2012. PMID: 23197831 Free PMC article.
-
Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum.PLoS Biol. 2009 Apr 14;7(4):e84. doi: 10.1371/journal.pbio.1000084. PLoS Biol. 2009. PMID: 19402747 Free PMC article.
-
CRISPR Interference of a Clonally Variant GC-Rich Noncoding RNA Family Leads to General Repression of var Genes in Plasmodium falciparum.mBio. 2020 Jan 21;11(1):e03054-19. doi: 10.1128/mBio.03054-19. mBio. 2020. PMID: 31964736 Free PMC article.
-
Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation.Trends Parasitol. 2008 Oct;24(10):455-61. doi: 10.1016/j.pt.2008.07.005. Epub 2008 Sep 2. Trends Parasitol. 2008. PMID: 18771955 Review.
-
A greedy promoter controls malarial var-iations.Cell. 2006 Jan 27;124(2):251-3. doi: 10.1016/j.cell.2006.01.004. Cell. 2006. PMID: 16439197 Review.
Cited by
-
Malaria Epigenetics.Cold Spring Harb Perspect Med. 2017 Jul 5;7(7):a025528. doi: 10.1101/cshperspect.a025528. Cold Spring Harb Perspect Med. 2017. PMID: 28320828 Free PMC article. Review.
-
Chromatin Accessibility-Based Characterization of the Gene Regulatory Network Underlying Plasmodium falciparum Blood-Stage Development.Cell Host Microbe. 2018 Apr 11;23(4):557-569.e9. doi: 10.1016/j.chom.2018.03.007. Cell Host Microbe. 2018. PMID: 29649445 Free PMC article.
-
Independent regulation of Plasmodium falciparum rif gene promoters.Sci Rep. 2018 Jun 19;8(1):9332. doi: 10.1038/s41598-018-27646-0. Sci Rep. 2018. PMID: 29921926 Free PMC article.
-
The Plasmodium falciparum histone methyltransferase PfSET10 is dispensable for the regulation of antigenic variation and gene expression in blood-stage parasites.mSphere. 2024 Nov 21;9(11):e0054624. doi: 10.1128/msphere.00546-24. Epub 2024 Oct 24. mSphere. 2024. PMID: 39445826 Free PMC article.
-
Rapid activation of distinct members of multigene families in Plasmodium spp.Commun Biol. 2020 Jul 3;3(1):351. doi: 10.1038/s42003-020-1081-3. Commun Biol. 2020. PMID: 32620892 Free PMC article.
References
-
- Beeson JG, Duffy PE. The immunology and pathogenesis of malaria during pregnancy. Curr Top Microbiol Immunol. 2005;297:187–227. - PubMed
-
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

