The intermediate-activity (L597V)BRAF mutant acts as an epistatic modifier of oncogenic RAS by enhancing signaling through the RAF/MEK/ERK pathway
- PMID: 22892241
- PMCID: PMC3435497
- DOI: 10.1101/gad.193458.112
The intermediate-activity (L597V)BRAF mutant acts as an epistatic modifier of oncogenic RAS by enhancing signaling through the RAF/MEK/ERK pathway
Abstract
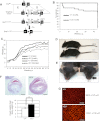
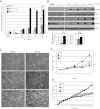
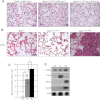
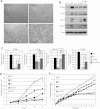
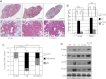
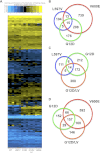
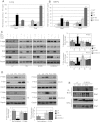
(L597V)BRAF mutations are acquired somatically in human cancer samples and are frequently coincident with RAS mutations. Germline (L597V)BRAF mutations are also found in several autosomal dominant developmental conditions known as RASopathies, raising the important question of how the same mutation can contribute to both pathologies. Using a conditional knock-in mouse model, we show that endogenous expression of (L597V)Braf leads to approximately twofold elevated Braf kinase activity and weak activation of the Mek/Erk pathway. This is associated with induction of RASopathy hallmarks including cardiac abnormalities and facial dysmorphia but is not sufficient for tumor formation. We combined (L597V)Braf with (G12D)Kras and found that (L597V)Braf modified (G12D)Kras oncogenesis such that fibroblast transformation and lung tumor development were more reminiscent of that driven by the high-activity (V600E)Braf mutant. Mek/Erk activation levels were comparable with those driven by (V600E)Braf in the double-mutant cells, and the gene expression signature was more similar to that induced by (V600E)Braf than (G12D)Kras. However, unlike (V600E)Braf, Mek/Erk pathway activation was mediated by both Craf and Braf, and ATP-competitive RAF inhibitors induced paradoxical Mek/Erk pathway activation. Our data show that weak activation of the Mek/Erk pathway underpins RASopathies, but in cancer, (L597V)Braf epistatically modifies the transforming effects of driver oncogenes.
Figures
References
-
- Ashworth A, Lord CJ, Reis-Filho JS 2011. Genetic interactions in cancer progression and treatment. Cell 145: 30–38 - PubMed
-
- Basso AD, Kirschmeier P, Bishop WR 2006. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res 47: 15–31 - PubMed
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
