A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons
- PMID: 22892276
- PMCID: PMC3514681
- DOI: 10.1101/gr.138651.112
A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons
Abstract
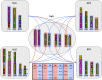
Chromosome rearrangements in small apes are up to 20 times more frequent than in most mammals. Because of their complexity, the full extent of chromosome evolution in these hominoids is not yet fully documented. However, previous work with array painting, BAC-FISH, and selective sequencing in two of the four karyomorphs has shown that high-resolution methods can precisely define chromosome breakpoints and map the complex flow of evolutionary chromosome rearrangements. Here we use these tools to precisely define the rearrangements that have occurred in the remaining two karyomorphs, genera Symphalangus (2n = 50) and Hoolock (2n = 38). This research provides the most comprehensive insight into the evolutionary origins of chromosome rearrangements involved in transforming small apes genome. Bioinformatics analyses of the human-gibbon synteny breakpoints revealed association with transposable elements and segmental duplications, providing some insight into the mechanisms that might have promoted rearrangements in small apes. In the near future, the comparison of gibbon genome sequences will provide novel insights to test hypotheses concerning the mechanisms of chromosome evolution. The precise definition of synteny block boundaries and orientation, chromosomal fusions, and centromere repositioning events presented here will facilitate genome sequence assembly for these close relatives of humans.
Figures




References
-
- Brown WR, Xu ZY 2009. The ‘kinetochore maintenance loop': The mark of regulation? Bioessays 31: 228–236 - PubMed
-
- Carbone L, D'Addabbo P, Cardone MF, Teti MG, Misceo D, Vessere GM, de Jong PJ, Rocchi M 2009a. A satellite-like sequence, representing a “clone gap” in the human genome, was likely involved in the seeding of a novel centromere in macaque. Chromosoma 118: 269–277 - PubMed
-
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J, et al. 2009b. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet 5: e1000538 doi: 10.1371/journal.pgen.1000538 - PMC - PubMed
-
- Chi JX, Huang L, Nie W, Wang J, Su B, Yang F 2005. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac hromosomes by BAC mapping. Chromosoma 114: 167–172 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
