The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice
- PMID: 22892321
- PMCID: PMC3462628
- DOI: 10.1105/tpc.112.101436
The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice
Abstract
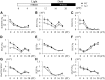
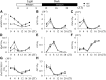
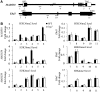
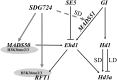
Chromatin modifications affect flowering time in the long-day plant Arabidopsis thaliana, but the role of histone methylation in flowering time regulation of rice (Oryza sativa), a short-day plant, remains to be elucidated. We identified a late-flowering long vegetative phase1 (lvp1) mutant in rice and used map-based cloning to reveal that lvp1 affects the SET domain group protein 724 (SDG724). SDG724 functions as a histone methyltransferase in vitro and contributes to a major fraction of global histone H3 lysine 36 (H3K36) methylation in vivo. Expression analyses of flowering time genes in wild-type and lvp1 mutants revealed that Early heading date1, but not Heading date1, are misregulated in lvp1 mutants. In addition, the double mutant of lvp1 with photoperiod sensitivity5 (se5) flowered later than the se5 single mutant, indicating that lvp1 delays flowering time irrespective of photoperiod. Chromatin immunoprecipitation assays showed that lvp1 had reduced levels of H3K36me2/3 at MADS50 and RFT1. This suggests that the divergent functions of paralogs RFT1 and Hd3a, and of MADS50 and MADS51, are in part due to differential H3K36me2/3 deposition, which also correlates with higher expression levels of MADS50 and RFT1 in flowering promotion in rice.
Figures




Similar articles
-
Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3.Plant Physiol. 2014 Mar;164(3):1326-37. doi: 10.1104/pp.113.228049. Epub 2014 Jan 13. Plant Physiol. 2014. PMID: 24420930 Free PMC article.
-
SET DOMAIN GROUP 708, a histone H3 lysine 36-specific methyltransferase, controls flowering time in rice (Oryza sativa).New Phytol. 2016 Apr;210(2):577-88. doi: 10.1111/nph.13768. Epub 2015 Dec 7. New Phytol. 2016. PMID: 26639303
-
The Rice CHD3/Mi-2 Chromatin Remodeling Factor Rolled Fine Striped Promotes Flowering Independent of Photoperiod.Int J Mol Sci. 2021 Jan 28;22(3):1303. doi: 10.3390/ijms22031303. Int J Mol Sci. 2021. PMID: 33525623 Free PMC article.
-
Genetic control of flowering time in rice: integration of Mendelian genetics and genomics.Theor Appl Genet. 2016 Dec;129(12):2241-2252. doi: 10.1007/s00122-016-2773-4. Epub 2016 Sep 30. Theor Appl Genet. 2016. PMID: 27695876 Review.
-
Environmental control of rice flowering time.Plant Commun. 2023 Sep 11;4(5):100610. doi: 10.1016/j.xplc.2023.100610. Epub 2023 May 4. Plant Commun. 2023. PMID: 37147799 Free PMC article. Review.
Cited by
-
A Green Light to Switch on Genes: Revisiting Trithorax on Plants.Plants (Basel). 2022 Dec 23;12(1):75. doi: 10.3390/plants12010075. Plants (Basel). 2022. PMID: 36616203 Free PMC article. Review.
-
Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3.Plant Physiol. 2014 Mar;164(3):1326-37. doi: 10.1104/pp.113.228049. Epub 2014 Jan 13. Plant Physiol. 2014. PMID: 24420930 Free PMC article.
-
The COMPASS-Like Complex Promotes Flowering and Panicle Branching in Rice.Plant Physiol. 2018 Apr;176(4):2761-2771. doi: 10.1104/pp.17.01749. Epub 2018 Feb 12. Plant Physiol. 2018. PMID: 29440594 Free PMC article.
-
Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process.Front Plant Sci. 2015 Aug 5;6:607. doi: 10.3389/fpls.2015.00607. eCollection 2015. Front Plant Sci. 2015. PMID: 26300904 Free PMC article.
-
Integrated transcriptomic, transcriptional factors, and protein interaction reveal the regulatory mechanisms of flowering time in rice (Oryza sativa L.).Transgenic Res. 2025 Apr 17;34(1):21. doi: 10.1007/s11248-025-00439-8. Transgenic Res. 2025. PMID: 40246762
References
-
- Berr A., Shafiq S., Shen W.H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809: 567–576 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

