HSPRO controls early Nicotiana attenuata seedling growth during interaction with the fungus Piriformospora indica
- PMID: 22892352
- PMCID: PMC3461566
- DOI: 10.1104/pp.112.203976
HSPRO controls early Nicotiana attenuata seedling growth during interaction with the fungus Piriformospora indica
Abstract
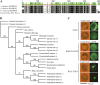
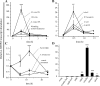
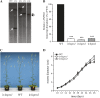
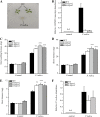
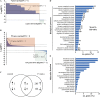
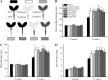
In a previous study aimed at identifying regulators of Nicotiana attenuata responses against chewing insects, a 26-nucleotide tag matching the HSPRO (ORTHOLOG OF SUGAR BEET Hs1(pro)(-)(1)) gene was found to be strongly induced after simulated herbivory (Gilardoni et al., 2010). Here we characterized the function of HSPRO during biotic interactions in transgenic N. attenuata plants silenced in its expression (ir-hspro). In wild-type plants, HSPRO expression was not only induced during simulated herbivory but also when leaves were inoculated with Pseudomonas syringae pv tomato DC3000 and roots with the growth-promoting fungus Piriformospora indica. Reduced HSPRO expression did not affect the regulation of direct defenses against Manduca sexta herbivory or P. syringae pv tomato DC3000 infection rates. However, reduced HSPRO expression positively influenced early seedling growth during interaction with P. indica; fungus-colonized ir-hspro seedlings increased their fresh biomass by 30% compared with the wild type. Grafting experiments demonstrated that reduced HSPRO expression in roots was sufficient to induce differential growth promotion in both roots and shoots. This effect was accompanied by changes in the expression of 417 genes in colonized roots, most of which were metabolic genes. The lack of major differences in the metabolic profiles of ir-hspro and wild-type colonized roots (as analyzed by liquid chromatography time-of-flight mass spectrometry) suggested that accelerated metabolic rates were involved. We conclude that HSPRO participates in a whole-plant change in growth physiology when seedlings interact with P. indica.
Figures
References
-
- Allmann S, Halitschke R, Schuurink RC, Baldwin IT. (2010) Oxylipin channelling in Nicotiana attenuata: lipoxygenase 2 supplies substrates for green leaf volatile production. Plant Cell Environ 33: 2028–2040 - PubMed
-
- Baldwin IT, Morse L. (1994) Up in smoke 2: germination of Nicotiana attenuata in response to smoke-derived cues and nutrients in burned and unburned soils. J Chem Ecol 20: 2373–2391 - PubMed
-
- Baldwin IT, Preston CA. (1999) The eco-physiological complexity of plant responses to insect herbivores. Planta 208: 137–145
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

