The Diversity Outbred mouse population
- PMID: 22892839
- PMCID: PMC3524832
- DOI: 10.1007/s00335-012-9414-2
The Diversity Outbred mouse population
Abstract
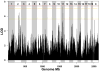
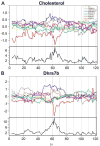
The Diversity Outbred (DO) population is a heterogeneous stock derived from the same eight founder strains as the Collaborative Cross (CC) inbred strains. Genetically heterogeneous DO mice display a broad range of phenotypes. Natural levels of heterozygosity provide genetic buffering and, as a result, DO mice are robust and breed well. Genetic mapping analysis in the DO presents new challenges and opportunities. Specialized algorithms are required to reconstruct haplotypes from high-density SNP array data. The eight founder haplotypes can be combined into 36 possible diplotypes, which must be accommodated in QTL mapping analysis. Population structure of the DO must be taken into account here. Estimated allele effects of eight founder haplotypes provide information that is not available in two-parent crosses and can dramatically reduce the number of candidate loci. Allele effects can also distinguish chance colocation of QTL from pleiotropy, which provides a basis for establishing causality in expression QTL studies. We recommended sample sizes of 200-800 mice for QTL mapping studies, larger than for traditional crosses. The CC inbred strains provide a resource for independent validation of DO mapping results. Genetic heterogeneity of the DO can provide a powerful advantage in our ability to generalize conclusions to other genetically diverse populations. Genetic diversity can also help to avoid the pitfall of identifying an idiosyncratic reaction that occurs only in a limited genetic context. Informatics tools and data resources associated with the CC, the DO, and their founder strains are developing rapidly. We anticipate a flood of new results to follow as our community begins to adopt and utilize these new genetic resource populations.
Figures
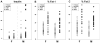
References
-
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19(6):382–389. - PMC - PubMed
-
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

