Computational models for large-scale simulations of facilitated diffusion
- PMID: 22892851
- PMCID: PMC4007627
- DOI: 10.1039/c2mb25201e
Computational models for large-scale simulations of facilitated diffusion
Abstract
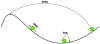
The binding of site-specific transcription factors to their genomic target sites is a key step in gene regulation. While the genome is huge, transcription factors belong to the least abundant protein classes in the cell. It is therefore fascinating how short the time frame is that they require to home in on their target sites. The underlying search mechanism is called facilitated diffusion and assumes a combination of three-dimensional diffusion in the space around the DNA combined with one-dimensional random walk on it. In this review, we present the current understanding of the facilitated diffusion mechanism and identify questions that lack a clear or detailed answer. One way to investigate these questions is through stochastic simulation and, in this manuscript, we support the idea that such simulations are able to address them. Finally, we review which biological parameters need to be included in such computational models in order to obtain a detailed representation of the actual process.
Figures
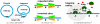
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

