Quantitative measurement of allele-specific protein expression in a diploid yeast hybrid by LC-MS
- PMID: 22893000
- PMCID: PMC3435501
- DOI: 10.1038/msb.2012.34
Quantitative measurement of allele-specific protein expression in a diploid yeast hybrid by LC-MS
Abstract
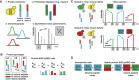
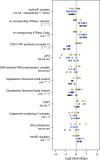
Understanding the genetic basis of gene regulatory variation is a key goal of evolutionary and medical genetics. Regulatory variation can act in an allele-specific manner (cis-acting) or it can affect both alleles of a gene (trans-acting). Differential allele-specific expression (ASE), in which the expression of one allele differs from another in a diploid, implies the presence of cis-acting regulatory variation. While microarrays and high-throughput sequencing have enabled genome-wide measurements of transcriptional ASE, methods for measurement of protein ASE (pASE) have lagged far behind. We describe a flexible, accurate, and scalable strategy for measurement of pASE by liquid chromatography-coupled mass spectrometry (LC-MS). We apply this approach to a hybrid between the yeast species Saccharomyces cerevisiae and Saccharomyces bayanus. Our results provide the first analysis of the relative contribution of cis-acting and trans-acting regulatory differences to protein expression divergence between yeast species.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

