TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export
- PMID: 22893130
- PMCID: PMC3654228
- DOI: 10.1038/ncomms2005
TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export
Abstract
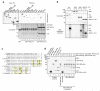
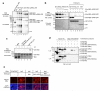
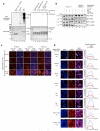
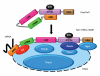
The metazoan TREX complex is recruited to mRNA during nuclear RNA processing and functions in exporting mRNA to the cytoplasm. Nxf1 is an mRNA export receptor, which binds processed mRNA and transports it through the nuclear pore complex. At present, the relationship between TREX and Nxf1 is not understood. Here we show that Nxf1 uses an intramolecular interaction to inhibit its own RNA-binding activity. When the TREX subunits Aly and Thoc5 make contact with Nxf1, Nxf1 is driven into an open conformation, exposing its RNA-binding domain, allowing RNA binding. Moreover, the combined knockdown of Aly and Thoc5 markedly reduces the amount of Nxf1 bound to mRNA in vivo and also causes a severe mRNA export block. Together, our data indicate that TREX provides a license for mRNA export by driving Nxf1 into a conformation capable of binding mRNA.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

