A novel approach to the discovery of small-molecule ligands of CDK2
- PMID: 22893598
- PMCID: PMC3483082
- DOI: 10.1002/cbic.201200316
A novel approach to the discovery of small-molecule ligands of CDK2
Abstract
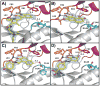
In an attempt to identify novel small-molecule ligands of cyclin-dependent kinase 2 (CDK2) with potential as allosteric inhibitors, we have devised a robust and cost-effective fluorescence-based high-throughput screening assay. The assay is based on the specific interaction of CDK2 with the extrinsic fluorophore 8-anilino-1-naphthalene sulfonate (ANS), which binds to a large allosteric pocket adjacent to the ATP site. Hit compounds that displace ANS directly or indirectly from CDK2 are readily classified as ATP site binders or allosteric ligands through the use of staurosporine, which blocks the ATP site without displacing ANS. Pilot screening of 1453 compounds led to the discovery of 12 compounds with displacement activities (EC(50) values) ranging from 6 to 44 μM, all of which were classified as ATP-site-directed ligands. Four new type I inhibitor scaffolds were confirmed by X-ray crystallography. Although this small compound library contained only ATP-site-directed ligands, the application of this assay to large compound libraries has the potential to reveal previously unrecognized chemical scaffolds suitable for structure-based design of CDK2 inhibitors with new mechanisms of action.
Copyright © 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


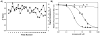

Similar articles
-
Discovery of a potential allosteric ligand binding site in CDK2.ACS Chem Biol. 2011 May 20;6(5):492-501. doi: 10.1021/cb100410m. Epub 2011 Feb 22. ACS Chem Biol. 2011. PMID: 21291269 Free PMC article.
-
Cooperativity Between Orthosteric Inhibitors and Allosteric Inhibitor 8-Anilino-1-Naphthalene Sulfonic Acid (ANS) in Cyclin-Dependent Kinase 2.ACS Chem Biol. 2020 Jul 17;15(7):1759-1764. doi: 10.1021/acschembio.0c00169. Epub 2020 May 20. ACS Chem Biol. 2020. PMID: 32433863 Free PMC article.
-
Structure-based discovery of the first allosteric inhibitors of cyclin-dependent kinase 2.Cell Cycle. 2014;13(14):2296-305. doi: 10.4161/cc.29295. Epub 2014 Jun 9. Cell Cycle. 2014. PMID: 24911186 Free PMC article.
-
Insights on Structural Characteristics and Ligand Binding Mechanisms of CDK2.Int J Mol Sci. 2015 Apr 24;16(5):9314-40. doi: 10.3390/ijms16059314. Int J Mol Sci. 2015. PMID: 25918937 Free PMC article. Review.
-
The rise of fragment-based drug discovery.Nat Chem. 2009 Jun;1(3):187-92. doi: 10.1038/nchem.217. Nat Chem. 2009. PMID: 21378847 Review.
Cited by
-
Virtual Screening for Potential Allosteric Inhibitors of Cyclin-Dependent Kinase 2 from Traditional Chinese Medicine.Molecules. 2016 Sep 21;21(9):1259. doi: 10.3390/molecules21091259. Molecules. 2016. PMID: 27657032 Free PMC article.
-
TWN-RENCOD: A novel method for protein binding site comparison.Comput Struct Biotechnol J. 2022 Dec 19;21:425-431. doi: 10.1016/j.csbj.2022.12.014. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36618985 Free PMC article.
-
A simple model-free method for direct assessment of fluorescent ligand binding by linear spectral summation.J Fluoresc. 2014 Jan;24(1):231-8. doi: 10.1007/s10895-013-1290-y. Epub 2013 Sep 18. J Fluoresc. 2014. PMID: 24043458 Free PMC article.
-
LifeSoaks: a tool for analyzing solvent channels in protein crystals and obstacles for soaking experiments.Acta Crystallogr D Struct Biol. 2023 Sep 1;79(Pt 9):837-856. doi: 10.1107/S205979832300582X. Epub 2023 Aug 10. Acta Crystallogr D Struct Biol. 2023. PMID: 37561404 Free PMC article.
-
Cyclin-Dependent Kinase Inhibitors as Anticancer Therapeutics.Mol Pharmacol. 2015 Nov;88(5):846-52. doi: 10.1124/mol.115.099325. Epub 2015 May 27. Mol Pharmacol. 2015. PMID: 26018905 Free PMC article.
References
-
- Johnson LN. Quarterly reviews of biophysics. 2009;42:1–40. - PubMed
- Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. - PubMed
- Zhang J, Yang PL, Gray NS. Nat Rev Cancer. 2009;9:28–39. - PubMed
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. Biochem J. 2007;408:297–315. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

