Probing spatial organization of DNA strands using enzyme-free hairpin assembly circuits
- PMID: 22894754
- PMCID: PMC3462741
- DOI: 10.1021/ja300984b
Probing spatial organization of DNA strands using enzyme-free hairpin assembly circuits
Abstract
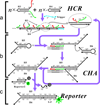
Catalyzed hairpin assembly (CHA) is a robust enzyme-free signal-amplification reaction that has a wide range of potential applications, especially in biosensing. Although most studies of the analytical applications of CHA have focused on the measurement of concentrations of biomolecules, we show here that CHA can also be used to probe the spatial organization of biomolecules such as single-stranded DNA. The basis of such detection is the fact that a DNA structure that brings a toehold and a branch-migration domain into close proximity can catalyze the CHA reaction. We quantitatively studied this phenomenon and applied it to the detection of domain reorganization that occurs during DNA self-assembly processes such as the hybridization chain reaction (HCR). We also show that CHA circuits can be designed to detect certain types of hybridization defects. This principle allowed us to develop a "signal on" assay that can simultaneously respond to multiple types of mutations in a DNA strand in one simple reaction, which is of great interest in genotyping and molecular diagnostics. These findings highlight the potential impacts of DNA circuitry on DNA nanotechnology and provide new tools for further development of these fields.
Figures


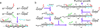


References
-
- Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. - PMC - PubMed
- Stojanovic MN, Landry DW. J. Am. Chem. Soc. 2002;124:9678–9679. - PubMed
- Cho EJ, Yang LT, Levy M, Ellington AD. J. Am. Chem. Soc. 2005;127:2022–2023. - PubMed
- Lubin AA, Plaxco KW. Accounts Chem. Res. 2010;43:496–505. - PMC - PubMed
- Stojanovic MN, Stefanovic D. Nat. Biotechnol. 2003;21:1069–1074. - PubMed
- Zhang L, Zhu J, Li T, Wang E. Anal. Chem. 2011;83:8871–8876. - PubMed
-
- Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. USA. 2004;101:15275–15278. - PMC - PubMed
- Qian L, Winfree E, Bruck J. Nature. 2011;475:368–372. - PubMed
- Zhang DY. Science. 2007;318:1121–1125. - PubMed
- Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. - PubMed
- Liu QH, Wang LM, Frutos AG, Condon AE, Corn RM, Smith LM. Nature. 2000;403:175–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

