Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1
- PMID: 22894777
- PMCID: PMC3420552
- DOI: 10.1089/hyb.2012.0009
Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1
Abstract
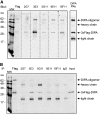
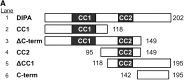
The coiled-coil domain-containing delta-interacting protein A (DIPA) is a transcription factor implicated in developmental regulation. DIPA is the first protein discovered to selectively interact with the p120-catenin (p120) isoform 1, an alternatively spliced form of p120 expressed preferentially in mesenchymal cells. Although a small fraction of p120 can be observed in the nucleus under certain circumstances, the vast majority of it associates with classical cadherins at adherens junctions. We observed for the first time that a discrete fraction of DIPA exists at cell-cell junctions, in addition to its predominantly nuclear localization. Thus, the p120-DIPA interaction may regulate cell signaling and/or transcriptional events, as has been described previously for β-catenin and the LEF/TCF transcription factor family. To facilitate further study of DIPA and to determine the physiological relevance of its interaction with p120, we have generated and characterized a panel of five DIPA-specific monoclonal antibodies (MAbs) that function in immunoblotting, immunoprecipitation, and immunofluorescence assays.
Figures






References
-
- Anastasiadis PZ. Moon SY. Thoreson MA. Mariner DJ. Crawford HC. Zheng Y. Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. - PubMed
-
- Wildenberg GA. Dohn MR. Carnahan RH. Davis MA. Lobdell NA. Settleman J. Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

