An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function
- PMID: 22894906
- PMCID: PMC3515020
- DOI: 10.4161/epi.21568
An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function
Abstract
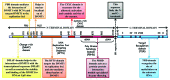
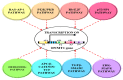
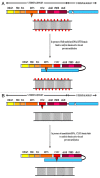
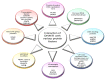
DNA methylation is one of the principal epigenetic signals that participate in cell specific gene expression in vertebrates. DNA methylation plays a quintessential role in the control of gene expression, cellular differentiation and development. It also plays a central role in the preservation of chromatin structure and chromosomal integrity, parental imprinting, X-chromosome inactivation, aging and carcinogenesis. The foremost contributor in the mammalian methylation scheme is DNMT1, a maintenance methyltransferase that faithfully copies the pre-existing methyl marks onto hemimethylated daughter strands during DNA replication to maintain the established methylation patterns across successive cell divisions. The ever-changing cellular physiology and the significant part that DNA methylation plays in genome regulation necessitate rigid management of this enzyme. In mammalian cells, a host of intrinsic and extrinsic mechanisms regulate the expression, activity and stability of DNMT1. Transcriptional regulation, post-transcriptional auto-inhibitory controls and post-translational modifications of the enzyme are responsible for the efficient inheritance of DNA methylation patterns. Also, a large number of intra- and intercellular signaling cascades and numerous interactions with other modulator molecules that affect the catalytic activity of the enzyme at multiple levels function as major checkpoints of the DNMT1 control system. An in-depth understanding of the DNMT1 enzyme, its targeting and function is crucial for comprehending how DNA methylation is coordinated with other critical developmental and physiological processes. This review aims to provide a comprehensive account of the various regulatory mechanisms and interactions of DNMT1 so as to elucidate its function at the molecular level and understand the dynamics of DNA methylation at the cellular level.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
