A novel image-based cytometry method for autophagy detection in living cells
- PMID: 22895056
- PMCID: PMC3442883
- DOI: 10.4161/auto.21028
A novel image-based cytometry method for autophagy detection in living cells
Abstract
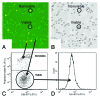
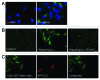
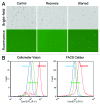
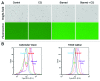
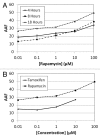
Autophagy is an important cellular catabolic process that plays a variety of important roles, including maintenance of the amino acid pool during starvation, recycling of damaged proteins and organelles, and clearance of intracellular microbes. Currently employed autophagy detection methods include fluorescence microscopy, biochemical measurement, SDS-PAGE and western blotting, but they are time consuming, labor intensive, and require much experience for accurate interpretation. More recently, development of novel fluorescent probes have allowed the investigation of autophagy via standard flow cytometry. However, flow cytometers remain relatively expensive and require a considerable amount of maintenance. Previously, image-based cytometry has been shown to perform automated fluorescence-based cellular analysis comparable to flow cytometry. In this study, we developed a novel method using the Cellometer image-based cytometer in combination with Cyto-ID(®) Green dye for autophagy detection in live cells. The method is compared with flow cytometry by measuring macroautophagy in nutrient-starved Jurkat cells. Results demonstrate similar trends of autophagic response, but different magnitude of fluorescence signal increases, which may arise from different analysis approaches characteristic of the two instrument platforms. The possibility of using this method for drug discovery applications is also demonstrated through the measurement of dose-response kinetics upon induction of autophagy with rapamycin and tamoxifen. The described image-based cytometry/fluorescent dye method should serve as a useful addition to the current arsenal of techniques available in support of autophagy-based drug discovery relating to various pathological disorders.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
