Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents
- PMID: 22895161
- PMCID: PMC3526177
- DOI: 10.1038/ismej.2012.81
Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents
Abstract
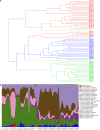
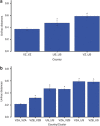
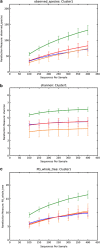
The human skin harbors complex bacterial communities. Prior studies showing high inter-individual variation focused on subjects from developed countries. We therefore compared cutaneous bacterial communities of Amerindians in the Venezuelan Amazon with subjects in the United States. Forearm skin specimens were studied from healthy Amerindians in Platanillal village in Amazonas State, and from healthy persons in New York and Colorado. All skin sampling used similar swab/buffer techniques. Multiplexed V2-targeted 16S rRNA gene pyrosequencing yielded high quality sequences from 112 samples. The results show 20 phyla, with three (Proteobacteria, Firmicutes, Actinobacteria) predominating. US residents and Venezuelan Amerindians had significantly different forearm skin bacterial community compositions, with United States dominated by Propionibacterium. Among the Amerindians, there was a deep split based on bacterial community membership, with 30 and 42 samples, respectively, falling into each of the two groups, not associated with age, gender, or body mass index. One Amerindian group had diversity similar to the United States, but was dominated by Staphylococcus rather than Propionibacterium. The other Amerindian group was significantly more diverse and even than the US or the other Amerindian group, and featured a broad range of Proteobacteria. The results provide evidence that ethnicity, lifestyle and/or geography are associated with the structure of human cutaneous bacterial communities.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

