Cistrome plasticity and mechanisms of cistrome reprogramming
- PMID: 22895178
- PMCID: PMC3466519
- DOI: 10.4161/cc.21281
Cistrome plasticity and mechanisms of cistrome reprogramming
Abstract
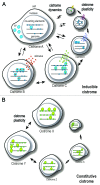
Mammalian genomes contain thousands of cis-regulatory elements for each transcription factor (TF), but TFs only occupy a relatively small subset referred to as cistrome. Recent studies demonstrate that a TF cistrome might differ among different organisms, tissue types and individuals. In a cell, a TF cistrome might differ among different physiological states, pathological stages and between physiological and pathological conditions. It is, therefore, remarkable how highly plastic these binding profiles are, and how massively they can be reprogrammed in rapid response to intra/extracellular variations and during cell identity transitions and evolution. Biologically, cistrome reprogramming events tend to be followed by changes in transcriptional outputs, thus serving as transformative mechanisms to synchronically alter the biology of the cell. In this review, we discuss the molecular basis of cistrome plasticity and attempt to integrate the different mechanisms and biological conditions associated with cistrome reprogramming. Emerging data suggest that, when altered, these reprogramming events might be linked to tumor development and/or progression, which is a radical conceptual change in our mechanistic understanding of cancer and, potentially, other diseases.
Figures



Similar articles
-
To bind or not to bind: Cistromic reprogramming in prostate cancer.Front Oncol. 2022 Sep 23;12:963007. doi: 10.3389/fonc.2022.963007. eCollection 2022. Front Oncol. 2022. PMID: 36212399 Free PMC article. Review.
-
Cistrome: an integrative platform for transcriptional regulation studies.Genome Biol. 2011 Aug 22;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. Genome Biol. 2011. PMID: 21859476 Free PMC article.
-
Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks.Nucleic Acids Res. 2019 Jul 2;47(W1):W206-W211. doi: 10.1093/nar/gkz332. Nucleic Acids Res. 2019. PMID: 31053864 Free PMC article.
-
Sea-ATI unravels novel vocabularies of plant active cistrome.Nucleic Acids Res. 2023 Nov 27;51(21):11568-11583. doi: 10.1093/nar/gkad853. Nucleic Acids Res. 2023. PMID: 37850650 Free PMC article.
-
Transcription networks rewire gene repertoire to coordinate cellular reprograming in prostate cancer.Semin Cancer Biol. 2023 Feb;89:76-91. doi: 10.1016/j.semcancer.2023.01.004. Epub 2023 Jan 23. Semin Cancer Biol. 2023. PMID: 36702449 Review.
Cited by
-
Limited conservation in cross-species comparison of GLK transcription factor binding suggested wide-spread cistrome divergence.Nat Commun. 2022 Dec 9;13(1):7632. doi: 10.1038/s41467-022-35438-4. Nat Commun. 2022. PMID: 36494366 Free PMC article.
-
The journey of neutropoiesis: how complex landscapes in bone marrow guide continuous neutrophil lineage determination.Blood. 2022 Apr 14;139(15):2285-2293. doi: 10.1182/blood.2021012835. Blood. 2022. PMID: 34986245 Free PMC article.
-
Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions.PLoS Genet. 2013 Nov;9(11):e1003906. doi: 10.1371/journal.pgen.1003906. Epub 2013 Nov 7. PLoS Genet. 2013. PMID: 24244184 Free PMC article.
-
Genome-wide activity of unliganded estrogen receptor-α in breast cancer cells.Proc Natl Acad Sci U S A. 2014 Apr 1;111(13):4892-7. doi: 10.1073/pnas.1315445111. Epub 2014 Mar 17. Proc Natl Acad Sci U S A. 2014. PMID: 24639548 Free PMC article.
-
Dissecting the genomic activity of a transcriptional regulator by the integrative analysis of omics data.Sci Rep. 2017 Aug 17;7(1):8564. doi: 10.1038/s41598-017-08754-9. Sci Rep. 2017. PMID: 28819152 Free PMC article.
References
-
- Gonzales KA, Ng HH. Choreographing pluripotency and cell fate with transcription factors. Biochim Biophys Acta 2011; 1809:337-49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
