Passenger deletions generate therapeutic vulnerabilities in cancer
- PMID: 22895339
- PMCID: PMC3712624
- DOI: 10.1038/nature11331
Passenger deletions generate therapeutic vulnerabilities in cancer
Erratum in
-
Corrigendum: Passenger deletions generate therapeutic vulnerabilities in cancer.Nature. 2015 Sep 10;525(7568):278. doi: 10.1038/nature14609. Epub 2015 Jul 8. Nature. 2015. PMID: 26153864 No abstract available.
Abstract
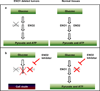
Inactivation of tumour-suppressor genes by homozygous deletion is a prototypic event in the cancer genome, yet such deletions often encompass neighbouring genes. We propose that homozygous deletions in such passenger genes can expose cancer-specific therapeutic vulnerabilities when the collaterally deleted gene is a member of a functionally redundant family of genes carrying out an essential function. The glycolytic gene enolase 1 (ENO1) in the 1p36 locus is deleted in glioblastoma (GBM), which is tolerated by the expression of ENO2. Here we show that short-hairpin-RNA-mediated silencing of ENO2 selectively inhibits growth, survival and the tumorigenic potential of ENO1-deleted GBM cells, and that the enolase inhibitor phosphonoacetohydroxamate is selectively toxic to ENO1-deleted GBM cells relative to ENO1-intact GBM cells or normal astrocytes. The principle of collateral vulnerability should be applicable to other passenger-deleted genes encoding functionally redundant essential activities and provide an effective treatment strategy for cancers containing such genomic events.
Figures



Comment in
-
Cancer: Exploiting collateral damage.Nature. 2012 Aug 16;488(7411):284-5. doi: 10.1038/488284a. Nature. 2012. PMID: 22895327 No abstract available.
-
Cancer: Passenger deletions create cancer-specific Achilles heel.Nat Rev Drug Discov. 2012 Sep;11(9):671. doi: 10.1038/nrd3826. Nat Rev Drug Discov. 2012. PMID: 22935798 No abstract available.
References
-
- Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. 10.1182/blood-2008-07-077958. - PubMed
-
- Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–374. 10.1038/nbt0511-373. - PubMed
-
- Weidle UH, Maisel D, Eick D. Synthetic lethality-based targets for discovery of new cancer therapeutics. Cancer Genomics Proteomics. 2011;8:159–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

