Koi herpesvirus encodes and expresses a functional interleukin-10
- PMID: 22896613
- PMCID: PMC3486318
- DOI: 10.1128/JVI.00957-12
Koi herpesvirus encodes and expresses a functional interleukin-10
Abstract
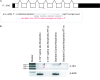
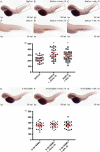
Koi herpesvirus (KHV) (species Cyprinid herpesvirus 3) ORF134 was shown to transcribe a spliced transcript encoding a 179-amino-acid (aa) interleukin-10 (IL-10) homolog (khvIL-10) in koi fin (KF-1) cells. Pairwise sequence alignment indicated that the expressed product shares 25% identity with carp IL-10, 22 to 24% identity with mammalian (including primate) IL-10s, and 19.1% identity with European eel herpesvirus IL-10 (ahvIL-10). In phylogenetic analyses, khvIL-10 fell in a divergent position from all host IL-10 sequences, indicating extensive structural divergence following capture from the host. In KHV-infected fish, khvIL-10 transcripts were observed to be highly expressed during the acute and reactivation phases but to be expressed at very low levels during low-temperature-induced persistence. Similarly, KHV early (helicase [Hel] and DNA polymerase [DNAP]) and late (intercapsomeric triplex protein [ITP] and major capsid protein [MCP]) genes were also expressed at high levels during the acute and reactivation phases, but only low-level expression of the ITP gene was detected during the persistent phase. Injection of khvIL-10 mRNA into zebrafish (Danio rerio) embryos increased the number of lysozyme-positive cells to a similar degree as zebrafish IL-10. Downregulation of the IL-10 receptor long chain (IL-10R1) using a specific morpholino abrogated the response to both khvIL-10 and zebrafish IL-10 transcripts, indicating that, despite the structural divergence, khvIL-10 functions via this receptor. This is the first report describing the characteristics of a functional viral IL-10 gene in the Alloherpesviridae.
Figures




Similar articles
-
Analysis of koi herpesvirus latency in wild common carp and ornamental koi in Oregon, USA.J Virol Methods. 2013 Feb;187(2):372-9. doi: 10.1016/j.jviromet.2012.11.015. Epub 2012 Nov 19. J Virol Methods. 2013. PMID: 23174162
-
Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae.J Gen Virol. 2005 Jun;86(Pt 6):1659-1667. doi: 10.1099/vir.0.80982-0. J Gen Virol. 2005. PMID: 15914843
-
Concentrations of a Koi herpesvirus (KHV) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR.Dis Aquat Organ. 2004 Sep 8;60(3):179-87. doi: 10.3354/dao060179. Dis Aquat Organ. 2004. PMID: 15521316
-
Koi herpesvirus (KHV) and KHV disease (KHVD) - a recently updated overview.J Appl Microbiol. 2020 Jul;129(1):98-103. doi: 10.1111/jam.14616. Epub 2020 Mar 8. J Appl Microbiol. 2020. PMID: 32077213 Review.
-
[Koi herpesvirus disease].Uirusu. 2005 Jun;55(1):145-51. doi: 10.2222/jsv.55.145. Uirusu. 2005. PMID: 16308541 Review. Japanese.
Cited by
-
Infectious hematopoietic necrosis virus (IHNV) persistence in Sockeye Salmon: influence on brain transcriptome and subsequent response to the viral mimic poly(I:C).BMC Genomics. 2015 Aug 26;16(1):634. doi: 10.1186/s12864-015-1759-y. BMC Genomics. 2015. PMID: 26306576 Free PMC article.
-
Street RABV Induces the Cholinergic Anti-inflammatory Pathway in Human Monocyte-Derived Macrophages by Binding to nAChr α7.Front Immunol. 2021 Feb 19;12:622516. doi: 10.3389/fimmu.2021.622516. eCollection 2021. Front Immunol. 2021. PMID: 33679766 Free PMC article.
-
The Function of Fish Cytokines.Biology (Basel). 2016 May 24;5(2):23. doi: 10.3390/biology5020023. Biology (Basel). 2016. PMID: 27231948 Free PMC article. Review.
-
Transcriptomic analysis of common carp anterior kidney during Cyprinid herpesvirus 3 infection: Immunoglobulin repertoire and homologue functional divergence.Sci Rep. 2017 Feb 2;7:41531. doi: 10.1038/srep41531. Sci Rep. 2017. PMID: 28148967 Free PMC article.
-
A tale of two fish: Comparative transcriptomics of resistant and susceptible steelhead following exposure to Ceratonova shasta highlights differences in parasite recognition.PLoS One. 2021 Feb 23;16(2):e0234837. doi: 10.1371/journal.pone.0234837. eCollection 2021. PLoS One. 2021. PMID: 33621237 Free PMC article.
References
-
- Aggad D, et al. 2010. In vivo analysis of IFN-γ1 and IFN-γ2 signaling in zebrafish. J. Immunol. 185:6774–6782 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Beck S, Barrell BG. 1988. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature 331:269–272 - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

