A three-helix junction is the interface between two functional domains of prohead RNA in 29 DNA packaging
- PMID: 22896620
- PMCID: PMC3486298
- DOI: 10.1128/JVI.01370-12
A three-helix junction is the interface between two functional domains of prohead RNA in 29 DNA packaging
Abstract
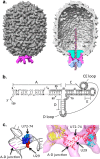
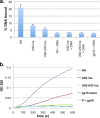
The double-stranded-DNA bacteriophages employ powerful molecular motors to translocate genomic DNA into preformed capsids during the packaging step in phage assembly. Bacillus subtilis bacteriophage 29 has an oligomeric prohead RNA (pRNA) that is an essential component of its packaging motor. The crystal structure of the pRNA-prohead binding domain suggested that a three-helix junction constitutes both a flexible region and part of a rigid RNA superhelix. Here we define the functional role of the three-helix junction in motor assembly and DNA packaging. Deletion mutagenesis showed that a U-rich region comprising two sides of the junction plays a role in the stable assembly of pRNA to the prohead. The retention of at least two bulged residues in this region was essential for pRNA binding and thereby subsequent DNA packaging. Additional deletions resulted in the loss of the ability of pRNA to multimerize in solution, consistent with the hypothesis that this region provides the flexibility required for pRNA oligomerization and prohead binding. The third side of the junction is part of a large RNA superhelix that spans the motor. The insertion of bases into this feature resulted in a loss of DNA packaging and an impairment of initiation complex assembly. Additionally, cryo-electron microscopy (cryoEM) analysis of third-side insertion mutants showed an increased flexibility of the helix that binds the ATPase, suggesting that the rigidity of the RNA superhelix is necessary for efficient motor assembly and function. These results highlight the critical role of the three-way junction in bridging the prohead binding and ATPase assembly functions of pRNA.
Figures



Similar articles
-
Prohead RNA: a noncoding viral RNA of novel structure and function.Wiley Interdiscip Rev RNA. 2016 Jul;7(4):428-37. doi: 10.1002/wrna.1330. Epub 2016 Jan 25. Wiley Interdiscip Rev RNA. 2016. PMID: 26810250 Free PMC article. Review.
-
An RNA Domain Imparts Specificity and Selectivity to a Viral DNA Packaging Motor.J Virol. 2015 Dec;89(24):12457-66. doi: 10.1128/JVI.01895-15. Epub 2015 Sep 30. J Virol. 2015. PMID: 26423956 Free PMC article.
-
Role of the CCA bulge of prohead RNA of bacteriophage ø29 in DNA packaging.J Mol Biol. 2008 Nov 14;383(3):520-8. doi: 10.1016/j.jmb.2008.08.056. Epub 2008 Aug 29. J Mol Biol. 2008. PMID: 18778713 Free PMC article.
-
Insights into the structure and assembly of the bacteriophage 29 double-stranded DNA packaging motor.J Virol. 2014 Apr;88(8):3986-96. doi: 10.1128/JVI.03203-13. Epub 2014 Jan 8. J Virol. 2014. PMID: 24403593 Free PMC article.
-
Current advances in Phi29 pRNA biology and its application in drug delivery.Wiley Interdiscip Rev RNA. 2012 Jul-Aug;3(4):469-81. doi: 10.1002/wrna.1111. Epub 2012 Feb 23. Wiley Interdiscip Rev RNA. 2012. PMID: 22362726 Review.
Cited by
-
Diverse self-association properties within a family of phage packaging RNAs.RNA. 2014 Nov;20(11):1759-74. doi: 10.1261/rna.045948.114. Epub 2014 Sep 22. RNA. 2014. PMID: 25246655 Free PMC article.
-
RNAloops: a database of RNA multiloops.Bioinformatics. 2022 Sep 2;38(17):4200-4205. doi: 10.1093/bioinformatics/btac484. Bioinformatics. 2022. PMID: 35809063 Free PMC article.
-
Prohead RNA: a noncoding viral RNA of novel structure and function.Wiley Interdiscip Rev RNA. 2016 Jul;7(4):428-37. doi: 10.1002/wrna.1330. Epub 2016 Jan 25. Wiley Interdiscip Rev RNA. 2016. PMID: 26810250 Free PMC article. Review.
-
Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA.RNA. 2017 Apr;23(4):521-529. doi: 10.1261/rna.059220.116. Epub 2017 Jan 9. RNA. 2017. PMID: 28069889 Free PMC article.
-
Three-way junction conformation dictates self-association of phage packaging RNAs.RNA Biol. 2016 Jul 2;13(7):635-45. doi: 10.1080/15476286.2016.1190075. Epub 2016 May 24. RNA Biol. 2016. PMID: 27217219 Free PMC article.
References
-
- Bailey S, et al. 1990. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J. Biol. Chem. 265:22365–22370 - PubMed
-
- Burroughs A, Iyer L, Aravind L. 2007. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems, p 48–65 In Volff J-N. (ed), Gene and protein evolution. Genomic dynamics, vol 3 Karger, Basel, Switzerland - PubMed
-
- Casjens SR. 2011. The DNA-packaging motor of tailed bacteriophages. Nat. Rev. Microbiol. 9:647–657 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

