Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage 6 procapsid
- PMID: 22896624
- PMCID: PMC3486324
- DOI: 10.1128/JVI.01347-12
Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage 6 procapsid
Abstract
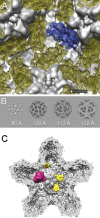
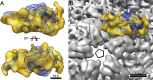
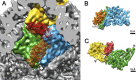
Bacteriophage 6 is a double-stranded RNA (dsRNA) virus whose genome is packaged sequentially as three single-stranded RNA (ssRNA) segments into an icosahedral procapsid which serves as a compartment for genome replication and transcription. The procapsid shell consists of 60 copies each of P1(A) and P1(B), two nonequivalent conformers of the P1 protein. Hexamers of the packaging ATPase P4 are mounted over the 5-fold vertices, and monomers of the RNA-dependent RNA polymerase (P2) attach to the inner surface, near the 3-fold axes. A fourth protein, P7, is needed for packaging and also promotes assembly. We used cryo-electron microscopy to localize P7 by difference mapping of procapsids with different protein compositions. We found that P7 resides on the interior surface of the P1 shell and appears to be monomeric. Its binding sites are arranged around the 3-fold axes, straddling the interface between two P1(A) subunits. Thus, P7 may promote assembly by stabilizing an initiation complex. Only about 20% of the 60 P7 binding sites were occupied in our preparations. P7 density overlaps P2 density similarly mapped, implying mutual occlusion. The known structure of the 12 homolog fits snugly into the P7 density. Both termini-which have been implicated in RNA binding-are oriented toward the adjacent 5-fold vertex, the entry pathway of ssRNA segments. Thus, P7 may promote packaging either by interacting directly with incoming RNA or by modulating the structure of the translocation pore.
Figures





Similar articles
-
Protein P7 of the cystovirus φ6 is located at the three-fold axis of the unexpanded procapsid.PLoS One. 2012;7(10):e47489. doi: 10.1371/journal.pone.0047489. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077625 Free PMC article.
-
Characterization of subunit-specific interactions in a double-stranded RNA virus: Raman difference spectroscopy of the phi6 procapsid.Biochemistry. 2002 Oct 8;41(40):11946-53. doi: 10.1021/bi0201623. Biochemistry. 2002. PMID: 12356294
-
Initial location of the RNA-dependent RNA polymerase in the bacteriophage Phi6 procapsid determined by cryo-electron microscopy.J Biol Chem. 2008 May 2;283(18):12227-31. doi: 10.1074/jbc.M710508200. Epub 2008 Feb 20. J Biol Chem. 2008. PMID: 18287088 Free PMC article.
-
Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives.Virus Res. 2004 Apr;101(1):83-92. doi: 10.1016/j.virusres.2003.12.008. Virus Res. 2004. PMID: 15010219 Review.
-
Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6.Microbiol Mol Biol Rev. 1999 Mar;63(1):149-60. doi: 10.1128/MMBR.63.1.149-160.1999. Microbiol Mol Biol Rev. 1999. PMID: 10066834 Free PMC article. Review.
Cited by
-
Single particle reconstruction and validation using Bsoft for the map challenge.J Struct Biol. 2018 Oct;204(1):90-95. doi: 10.1016/j.jsb.2018.07.003. Epub 2018 Jul 5. J Struct Biol. 2018. PMID: 29981840 Free PMC article.
-
Controlled Disassembly and Purification of Functional Viral Subassemblies Using Asymmetrical Flow Field-Flow Fractionation (AF4).Viruses. 2018 Oct 23;10(11):579. doi: 10.3390/v10110579. Viruses. 2018. PMID: 30360510 Free PMC article.
-
Cystoviral RNA-directed RNA polymerases: Regulation of RNA synthesis on multiple time and length scales.Virus Res. 2017 Apr 15;234:135-152. doi: 10.1016/j.virusres.2017.01.006. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104452 Free PMC article. Review.
-
Utilization of Bacteriophage phi6 for the Production of High-Quality Double-Stranded RNA Molecules.Viruses. 2024 Jan 22;16(1):166. doi: 10.3390/v16010166. Viruses. 2024. PMID: 38275976 Free PMC article. Review.
-
Cystoviral polymerase complex protein P7 uses its acidic C-terminal tail to regulate the RNA-directed RNA polymerase P2.J Mol Biol. 2014 Jul 15;426(14):2580-93. doi: 10.1016/j.jmb.2014.04.028. Epub 2014 May 9. J Mol Biol. 2014. PMID: 24813120 Free PMC article.
References
-
- Atherton BA, Cunningham EL, Splittgerber AG. 1996. A mathematical model for the description of the Coomassie brilliant blue protein assay. Anal. Biochem. 233:160–168 - PubMed
-
- Bamford JK, Bamford DH, Li T, Thomas GJ., Jr 1993. Structural studies of the enveloped dsRNA bacteriophage phi 6 of Pseudomonas syringae by Raman spectroscopy. II. Nucleocapsid structure and thermostability of the virion, nucleocapsid and polymerase complex. J. Mol. Biol. 230:473–482 - PubMed
-
- de Haas F, Paatero AO, Mindich L, Bamford DH, Fuller SD. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J. Mol. Biol. 294:357–372 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

