Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family
- PMID: 22896626
- PMCID: PMC3486290
- DOI: 10.1128/JVI.01023-12
Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family
Abstract
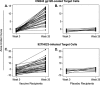
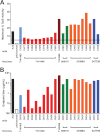
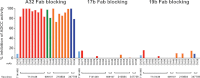
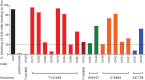
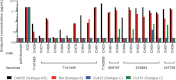
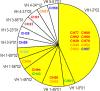
The ALVAC-HIV/AIDSVAX-B/E RV144 vaccine trial showed an estimated efficacy of 31%. RV144 secondary immune correlate analysis demonstrated that the combination of low plasma anti-HIV-1 Env IgA antibodies and high levels of antibody-dependent cellular cytotoxicity (ADCC) inversely correlate with infection risk. One hypothesis is that the observed protection in RV144 is partially due to ADCC-mediating antibodies. We found that the majority (73 to 90%) of a representative group of vaccinees displayed plasma ADCC activity, usually (96.2%) blocked by competition with the C1 region-specific A32 Fab fragment. Using memory B-cell cultures and antigen-specific B-cell sorting, we isolated 23 ADCC-mediating nonclonally related antibodies from 6 vaccine recipients. These antibodies targeted A32-blockable conformational epitopes (n = 19), a non-A32-blockable conformational epitope (n = 1), and the gp120 Env variable loops (n = 3). Fourteen antibodies mediated cross-clade target cell killing. ADCC-mediating antibodies displayed modest levels of V-heavy (VH) chain somatic mutation (0.5 to 1.5%) and also displayed a disproportionate usage of VH1 family genes (74%), a phenomenon recently described for CD4-binding site broadly neutralizing antibodies (bNAbs). Maximal ADCC activity of VH1 antibodies correlated with mutation frequency. The polyclonality and low mutation frequency of these VH1 antibodies reveal fundamental differences in the regulation and maturation of these ADCC-mediating responses compared to VH1 bNAbs.
Figures


Similar articles
-
Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.mBio. 2020 Jun 30;11(3):e00208-20. doi: 10.1128/mBio.00208-20. mBio. 2020. PMID: 32605979 Free PMC article.
-
HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities.J Virol. 2014 Jul;88(14):7715-26. doi: 10.1128/JVI.00156-14. Epub 2014 May 7. J Virol. 2014. PMID: 24807721 Free PMC article. Clinical Trial.
-
Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection.J Virol. 2014 Nov;88(21):12895-906. doi: 10.1128/JVI.02194-14. Epub 2014 Aug 27. J Virol. 2014. PMID: 25165110 Free PMC article.
-
Epitope specificity of human immunodeficiency virus-1 antibody dependent cellular cytotoxicity [ADCC] responses.Curr HIV Res. 2013 Jul;11(5):378-87. doi: 10.2174/1570162x113116660059. Curr HIV Res. 2013. PMID: 24191939 Free PMC article. Review.
-
HIV-specific antibody-dependent cellular cytotoxicity: a novel vaccine modality.Expert Rev Clin Immunol. 2012 Nov;8(8):767-74. doi: 10.1586/eci.12.74. Expert Rev Clin Immunol. 2012. PMID: 23167688 Review.
Cited by
-
A High Throughput Protein Microarray Approach to Classify HIV Monoclonal Antibodies and Variant Antigens.PLoS One. 2015 May 4;10(5):e0125581. doi: 10.1371/journal.pone.0125581. eCollection 2015. PLoS One. 2015. PMID: 25938510 Free PMC article.
-
Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.PLoS Pathog. 2016 Aug 25;12(8):e1005815. doi: 10.1371/journal.ppat.1005815. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27560183 Free PMC article.
-
Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization.J Immunol. 2015 Jun 15;194(12):5903-14. doi: 10.4049/jimmunol.1500122. Epub 2015 May 11. J Immunol. 2015. PMID: 25964491 Free PMC article.
-
Rationally Designed Immunogens Targeting HIV-1 gp120 V1V2 Induce Distinct Conformation-Specific Antibody Responses in Rabbits.J Virol. 2016 Nov 28;90(24):11007-11019. doi: 10.1128/JVI.01409-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707920 Free PMC article.
-
Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.J Virol. 2020 Jan 31;94(4):e01120-19. doi: 10.1128/JVI.01120-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776278 Free PMC article. Clinical Trial.
References
-
- Baum LL, et al. 1996. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 157:2168–2173 - PubMed
-
- Breden F, et al. 2011. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One 6:e16857 doi:10.1371/journal.pone.0016857 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

