Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner
- PMID: 22896699
- PMCID: PMC3464543
- DOI: 10.1074/jbc.M111.325332
Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner
Abstract
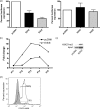
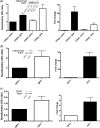
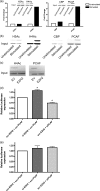
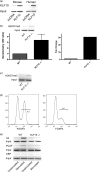
Inducible gene expression underlies the epigenetically inherited differentiation program of most immune cells. We report that the promoter of the FOXP3 gene possesses two distinct functional states: an "off state" mediated by the polycomb histone methyltransferase complex and a histone acetyltransferase-dependent "on state." Regulating these states is the presence of a Kruppel-like factor (KLF)-containing Polycomb response element. In the KLF10(-/-) mouse, the FOXP3 promoter is epigenetically silenced by EZH2 (Enhancer of Zeste 2)-mediated trimethylation of Histone 3 K27; thus, impaired FOXP3 induction and inappropriate adaptive T regulatory cell differentiation results in vitro and in vivo. The epigenetic transmittance of adaptive T regulatory cell deficiency is demonstrated throughout more than 40 generations of mice. These results provide insight into chromatin remodeling events key to phenotypic features of distinct T cell populations.
Figures



Similar articles
-
Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene.Am J Physiol Regul Integr Comp Physiol. 2014 Sep 15;307(6):R608-20. doi: 10.1152/ajpregu.00085.2014. Epub 2014 Jun 18. Am J Physiol Regul Integr Comp Physiol. 2014. PMID: 24944246 Free PMC article.
-
Disruption of FOXP3-EZH2 Interaction Represents a Pathobiological Mechanism in Intestinal Inflammation.Cell Mol Gastroenterol Hepatol. 2018 Sep 14;7(1):55-71. doi: 10.1016/j.jcmgh.2018.08.009. eCollection 2019. Cell Mol Gastroenterol Hepatol. 2018. PMID: 30510991 Free PMC article.
-
Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells.Nat Immunol. 2014 Jun;15(6):580-587. doi: 10.1038/ni.2868. Epub 2014 Apr 13. Nat Immunol. 2014. PMID: 24728351 Free PMC article.
-
Downregulation of Ezh2 methyltransferase by FOXP3: new insight of FOXP3 into chromatin remodeling?Biochim Biophys Acta. 2013 Oct;1833(10):2190-200. doi: 10.1016/j.bbamcr.2013.05.014. Epub 2013 May 17. Biochim Biophys Acta. 2013. PMID: 23688634
-
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2.Genes (Basel). 2023 Sep 27;14(10):1879. doi: 10.3390/genes14101879. Genes (Basel). 2023. PMID: 37895228 Free PMC article. Review.
Cited by
-
KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling.Cancers (Basel). 2018 May 25;10(6):161. doi: 10.3390/cancers10060161. Cancers (Basel). 2018. PMID: 29799499 Free PMC article. Review.
-
Androgens regulate ovarian gene expression by balancing Ezh2-Jmjd3 mediated H3K27me3 dynamics.PLoS Genet. 2021 Mar 30;17(3):e1009483. doi: 10.1371/journal.pgen.1009483. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33784295 Free PMC article.
-
Krüppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation.Am J Physiol Gastrointest Liver Physiol. 2015 Dec 1;309(11):G900-9. doi: 10.1152/ajpgi.00309.2015. Epub 2015 Oct 15. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 26472224 Free PMC article.
-
Epigenetics of Inflammatory Bowel Diseases.Turk J Gastroenterol. 2023 May;34(5):437-448. doi: 10.5152/tjg.2023.22515. Turk J Gastroenterol. 2023. PMID: 37158530 Free PMC article.
-
KLF2 is a rate-limiting transcription factor that can be targeted to enhance regulatory T-cell production.Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):9579-84. doi: 10.1073/pnas.1323493111. Epub 2014 Jun 16. Proc Natl Acad Sci U S A. 2014. PMID: 24979767 Free PMC article.
References
-
- Curotto de Lafaille M. A., Lafaille J. J. (2009) Natural and adaptive foxp3+ regulatory T cells. More of the same or a division of labor? Immunity 30, 626–635 - PubMed
-
- Li M. O., Sanjabi S., Flavell R. A. (2006) Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity 25, 455–471 - PubMed
-
- Li M. O., Wan Y. Y., Flavell R. A. (2007) T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 26, 579–591 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

