In silico directed mutagenesis identifies the CD81/claudin-1 hepatitis C virus receptor interface
- PMID: 22897233
- PMCID: PMC3549482
- DOI: 10.1111/cmi.12008
In silico directed mutagenesis identifies the CD81/claudin-1 hepatitis C virus receptor interface
Abstract
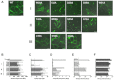
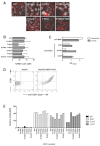
Hepatitis C virus (HCV) entry is dependent on host cell molecules tetraspanin CD81, scavenger receptor BI and tight junction proteins claudin-1 and occludin. We previously reported a role for CD81/claudin-1 receptor complexes in HCV entry; however, the molecular mechanism(s) driving association between the receptors is unknown. We explored the molecular interface between CD81 and claudin-1 using a combination of bioinformatic sequence-based modelling, site-directed mutagenesis and Fluorescent Resonance Energy Transfer (FRET) imaging methodologies. Structural modelling predicts the first extracellular loop of claudin-1 to have a flexible beta conformation and identifies a motif between amino acids 62-66 that interacts with CD81 residues T149, E152 and T153. FRET studies confirm a role for these CD81 residues in claudin-1 association and HCV infection. Importantly, mutation of these CD81 residues has minimal impact on protein conformation or HCV glycoprotein binding, highlighting a new functional domain of CD81 that is essential for virus entry.
© 2012 Blackwell Publishing Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

