Excited state dynamics in photosynthetic reaction center and light harvesting complex 1
- PMID: 22897312
- PMCID: PMC3427344
- DOI: 10.1063/1.4738953
Excited state dynamics in photosynthetic reaction center and light harvesting complex 1
Abstract
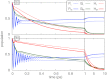
Key to efficient harvesting of sunlight in photosynthesis is the first energy conversion process in which electronic excitation establishes a trans-membrane charge gradient. This conversion is accomplished by the photosynthetic reaction center (RC) that is, in case of the purple photosynthetic bacterium Rhodobacter sphaeroides studied here, surrounded by light harvesting complex 1 (LH1). The RC employs six pigment molecules to initiate the conversion: four bacteriochlorophylls and two bacteriopheophytins. The excited states of these pigments interact very strongly and are simultaneously influenced by the surrounding thermal protein environment. Likewise, LH1 employs 32 bacteriochlorophylls influenced in their excited state dynamics by strong interaction between the pigments and by interaction with the protein environment. Modeling the excited state dynamics in the RC as well as in LH1 requires theoretical methods, which account for both pigment-pigment interaction and pigment-environment interaction. In the present study we describe the excitation dynamics within a RC and excitation transfer between light harvesting complex 1 (LH1) and RC, employing the hierarchical equation of motion method. For this purpose a set of model parameters that reproduce RC as well as LH1 spectra and observed oscillatory excitation dynamics in the RC is suggested. We find that the environment has a significant effect on LH1-RC excitation transfer and that excitation transfers incoherently between LH1 and RC.
Figures




References
-
- Blankenship R. E., Molecular Mechanisms of Photosynthesis (Blackwell Science, Malden, MA, 2002).
-
- Jonas D. M., Lang M. J., Nagasawa Y., Joo T., and Fleming G. R., “Pump-probe polarization anisotropy study of femtosecond energy transfer within the photosynthetic reaction center of rhodobacter sphaeroides r26,” J. Phys. Chem. 100, 12660–12673 (1996). 10.1021/jp960708t - DOI
-
- Cherepy N. J., Shreve A. P., Moore L. J., Boxer S. G., and Mathies R. A., “Temperature dependence of the qy resonance raman spectra of bacteriochlorophylls, the primary electron donor, and bacteriopheophytins in the bacterial photosynthetic reaction center,” Biochemistry 36(28), 8559–8566 (1997). 10.1021/bi970024r - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

