Targeting oncogenic Ras signaling in hematologic malignancies
- PMID: 22898602
- PMCID: PMC3482854
- DOI: 10.1182/blood-2012-05-378596
Targeting oncogenic Ras signaling in hematologic malignancies
Abstract
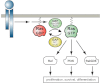
Ras proteins are critical nodes in cellular signaling that integrate inputs from activated cell surface receptors and other stimuli to modulate cell fate through a complex network of effector pathways. Oncogenic RAS mutations are found in ∼25% of human cancers and are highly prevalent in hematopoietic malignancies. Because of their structural and biochemical properties, oncogenic Ras proteins are exceedingly difficult targets for rational drug discovery, and no mechanism-based therapies exist for cancers with RAS mutations. This article reviews the properties of normal and oncogenic Ras proteins, the prevalence and likely pathogenic role of NRAS, KRAS, and NF1 mutations in hematopoietic malignancies, relevant animal models of these cancers, and implications for drug discovery. Because hematologic malignancies are experimentally tractable, they are especially valuable platforms for addressing the fundamental question of how to reverse the adverse biochemical output of oncogenic Ras in cancer.
Figures

References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. - PubMed
-
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. - PubMed
-
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–465. - PubMed
-
- Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10(12):1911–1918. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

