Bioactive natural products from Lysobacter
- PMID: 22898908
- PMCID: PMC3468324
- DOI: 10.1039/c2np20064c
Bioactive natural products from Lysobacter
Abstract
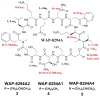
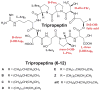
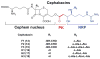
The gliding Gram-negative Lysobacter bacteria are emerging as a promising source of new bioactive natural products. These ubiquitous freshwater and soil microorganisms are fast growing, simple to use and maintain, and genetically amenable for biosynthetic engineering. This Highlight reviews a group of biologically active and structurally distinct natural products from the genus Lysobacter, with a focus on their biosyntheses. Although Lysobacter sp. are known as prolific producers of bioactive natural products, detailed molecular mechanistic studies of their enzymatic assembly have been surprisingly scarce. We hope to provide a snapshot of the important work done on the lysobacterial natural products and to provide useful information for future biosynthetic engineering of novel antibiotics in Lysobacter.
Figures



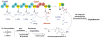



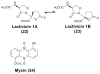
References
-
- Christensen P, Cook FD. Int J Syst Bacteriol. 1978;28:367–393.
-
- Reichenbach H. In: Prokaryotes. Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. Springer; New York: 2006. pp. 939–957.
-
- Sullivan RF, Holtman MA, Zylstra GJ, White JF, Kobayashi DY. J Appl Microbiol. 2003;94:1079–1086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

