A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation
- PMID: 22899282
- PMCID: PMC3514664
- DOI: 10.1101/gr.132738.111
A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation
Abstract
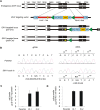
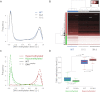
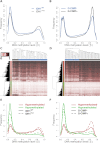
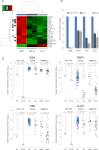
Monoallelic point mutations of the NADP(+)-dependent isocitrate dehydrogenases IDH1 and IDH2 occur frequently in gliomas, acute myeloid leukemias, and chondromas, and display robust association with specific DNA hypermethylation signatures. Here we show that heterozygous expression of the IDH1(R132H) allele is sufficient to induce the genome-wide alterations in DNA methylation characteristic of these tumors. Using a gene-targeting approach, we knocked-in a single copy of the most frequently observed IDH1 mutation, R132H, into a human cancer cell line and profiled changes in DNA methylation at over 27,000 CpG dinucleotides relative to wild-type parental cells. We find that IDH1(R132H/WT) mutation induces widespread alterations in DNA methylation, including hypermethylation of 2010 and hypomethylation of 842 CpG loci. We demonstrate that many of these alterations are consistent with those observed in IDH1-mutant and G-CIMP+ primary gliomas and can segregate IDH wild-type and mutated tumors as well as those exhibiting the G-CIMP phenotype in unsupervised analysis of two primary glioma cohorts. Further, we show that the direction of IDH1(R132H/WT)-mediated DNA methylation change is largely dependent upon preexisting DNA methylation levels, resulting in depletion of moderately methylated loci. Additionally, whereas the levels of multiple histone H3 and H4 methylation modifications were globally increased, consistent with broad inhibition of histone demethylation, hypermethylation at H3K9 in particular accompanied locus-specific DNA hypermethylation at several genes down-regulated in IDH1(R132H/WT) knock-in cells. These data provide insight on epigenetic alterations induced by IDH1 mutations and support a causal role for IDH1(R132H/WT) mutants in driving epigenetic instability in human cancer cells.
Figures



References
-
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 - PubMed
-
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116: 597–602 - PubMed
-
- Barbus S, Tews B, Karra D, Hahn M, Radlwimmer B, Delhomme N, Hartmann C, Felsberg J, Krex D, Schackert G, et al. 2011. Differential retinoic acid signaling in tumors of long- and short-term glioblastoma survivors. J Natl Cancer Inst 103: 598–606 - PubMed
-
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, et al. 2009. IDH1 mutations at residue p.R132 (IDH1R132) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat 30: 7–11 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
