Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica
- PMID: 22899817
- PMCID: PMC3507640
- DOI: 10.1186/1471-2164-13-394
Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica
Abstract
Background: Genomic imprinting is an epigenetic phenomenon resulting in parent-of-origin specific monoallelic gene expression. It is postulated to have evolved in placental mammals to modulate intrauterine resource allocation to the offspring. In this study, we determined the imprint status of metatherian orthologues of eutherian imprinted genes.
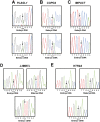
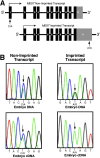
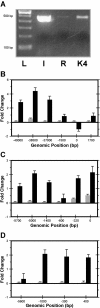
Results: L3MBTL and HTR2A were shown to be imprinted in Monodelphis domestica (the gray short-tailed opossum). MEST expressed a monoallelic and a biallelic transcript, as in eutherians. In contrast, IMPACT, COPG2, and PLAGL1 were not imprinted in the opossum. Differentially methylated regions (DMRs) involved in regulating imprinting in eutherians were not found at any of the new imprinted loci in the opossum. Interestingly, a novel DMR was identified in intron 11 of the imprinted IGF2R gene, but this was not conserved in eutherians. The promoter regions of the imprinted genes in the opossum were enriched for the activating histone modification H3 Lysine 4 dimethylation.
Conclusions: The phenomenon of genomic imprinting is conserved in Therians, but the marked difference in the number and location of imprinted genes and DMRs between metatherians and eutherians indicates that imprinting is not fully conserved between the two Therian infra-classes. The identification of a novel DMR at a non-conserved location as well as the first demonstration of histone modifications at imprinted loci in the opossum suggest that genomic imprinting may have evolved in a common ancestor of these two Therian infra-classes with subsequent divergence of regulatory mechanisms in the two lineages.
Figures


References
-
- McGrath J, Solter D. Nucleocytoplasmic interactions in the mouse embryo. J Embryol Exp Morphol. 1986;97(Suppl):277–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

